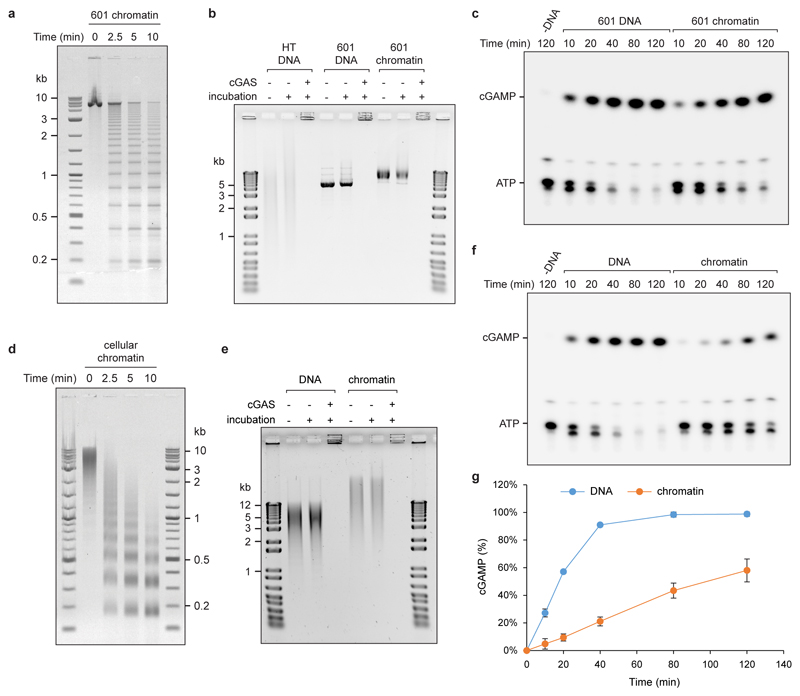
Extended data Fig 5. cGAS is activated by chromatin.
(a) Agarose gel of micrococcal nuclease (MNase) digested synthetic chromatin assembled onto a ‘601’ DNA template indicates it has a regular nucleosomal structure. (b) Chromatin and DNA bind recombinant cGAS; DNA in wells could be the result of near charge neutrality of cGAS-DNA complexes or previously reported cGAS oligomerisation. Chromatin is stable under cGAS assay conditions remaining intact during incubation in cGAS reaction buffer, as evidenced by the bandshift compared to naked DNA. (c) Representative TLC image demonstrating cGAMP generation by recombinant cGAS in the presence of chromatin. (d) MNase treatment confirms a nucleosomal ladder pattern for chromatin isolated from mouse NIH3T3 cells. (e) cGAS binds chromatin, and cellular chromatin is stable under cGAS assay conditions. (f, g) Cellular chromatin activates recombinant cGAS, but at a slower rate than the same amount of deproteinised DNA. Representative images shown. Graphs shows quantification from n=3 independent experiments, mean ± SD. Reduced cGAS activation in vitro by chromatin isolated from cells is expected due to 1) the presence of linker histones in addition to the nucleosomal core histones, which has been shown to bind part of the linker DNA, reducing the available sites for cGAS binding, and 2) the use of MNase during the isolation of cellular chromatin. Whereas MNase treatment is needed to fragment the chromatin to allow its purification it will preferentially cleave accessible non-protein-bound portions, which will further reduce the available sites to which cGAS can bind in the final chromatin preparation. However, such nucleosome-free regions are more likely to allow efficient binding and activation of cGAS in vivo.