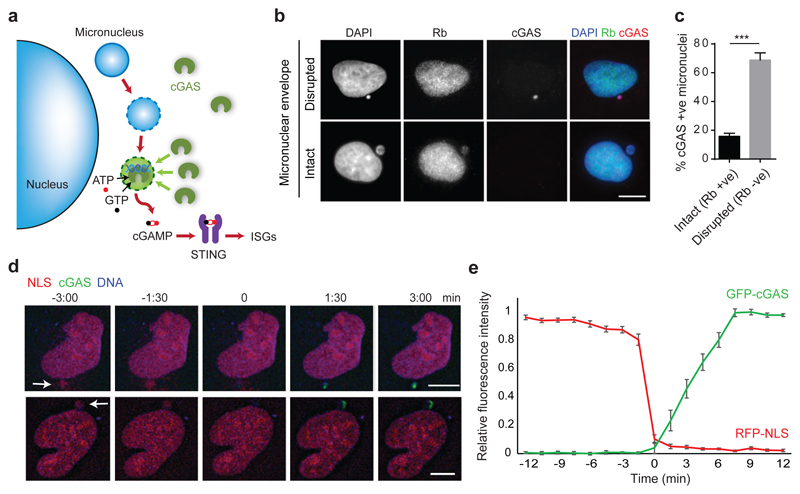
Fig 2. cGAS localises to micronuclei upon nuclear envelope rupture.
(a) Model: Micronuclear membrane rupture leads to cGAS sensing of DNA. Micronuclei are susceptible to nuclear envelope collapse, which permits cytosolic cGAS access to genomic dsDNA, initiating a cGAS-STING dependent proinflammatory immune response through production of the second messenger cGAMP. (b, c) cGAS localisation to micronuclei in U2OS cells inversely correlates with localisation of Rb, the latter present only in micronuclei with an intact nuclear envelope. (b) Representative images. (c) Quantification. Mean ± SEM of n=3 independent experiments (≥250 micronuclei counted per experiment); cGAS +ve : cGAS-stained micronuclei; Rb +ve/-ve: micronuclei positive/negative for Rb staining. (d) Representative stills from live imaging of U2OS cells expressing mCherry-NLS and GFP-cGAS. DNA visualised with Hoechst. Time (min) relative to loss of mCherry-NLS from micronucleus (t=0, micronuclear membrane rupture). Arrows indicate micronuclei undergoing rupture. (e) Quantification of cGAS signal accumulating in micronuclei after loss of nuclear envelope integrity. Relative mean fluorescence intensity plotted. Error bars, SEM of n=11 micronuclei.