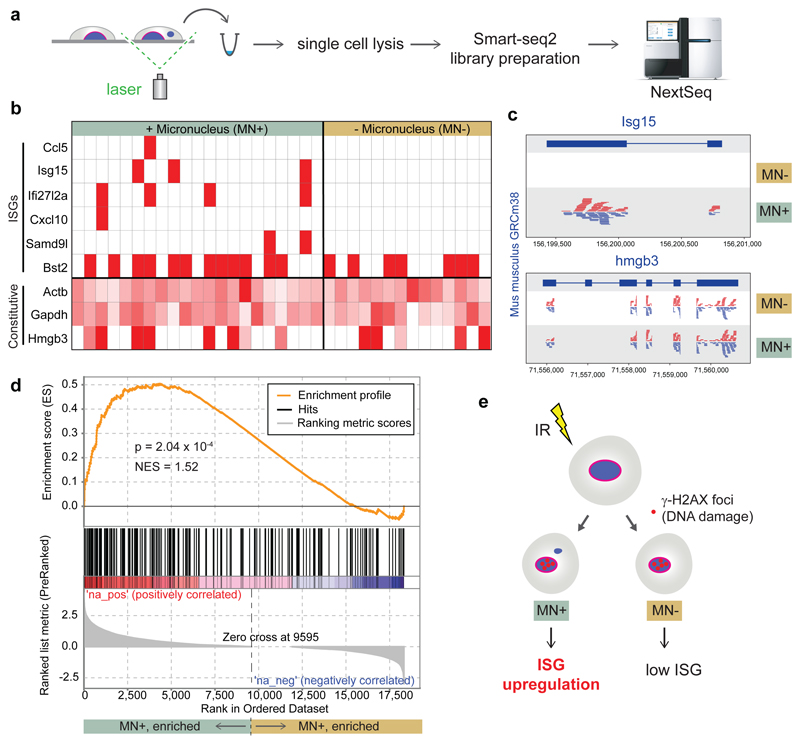
Fig 5. ISG upregulation occurs specifically in micronucleated cells following DNA damage.
(a) Experimental outline: 48 h after irradiation (1 Gy) of C57BL/6J MEFs, individual live cells with normal nuclear morphology (MN-) or with micronuclei (MN+) were identified microscopically and excised by laser microdissection. Single cell transcriptomes were then generated. (b, c) Transcripts of multiple ISGs were detected only in micronucleated cells. (b) Heat map for individual cells, with ISGs and constitutively expressed control genes (Actb, Gapdh, Hmgb3) indicated. Red boxes denote detection of ≥1 read per ISG. Control genes shaded in red according to read count relative to the maximum observed for each gene. (c) Alignment of pooled sequence reads to Isg15 and Hmg3b. (d) Transcriptome-wide analysis (GSEA) demonstrates that a set of 336 ISG genes is significantly enriched (P = 2.04 x 10-4) in transcriptomes of micronucleated cells. (e) Interpretation of single cell data: After induction of DNA damage by IR, ISG upregulation preferentially occurs in micronucleated cells to those with normal nuclear morphology exposed to identical experimental conditions.