Abstract
Two studies reveal that early-life malfunction in organelles called mitochondria brings about lasting changes in how DNA is packaged. These alterations have consequences for cellular stress responses and organismal longevity.
Organelles called mitochondria are central to the generation of ATP molecules, which are the energy currency of cells. Mild mitochondrial impairment in juvenile roundworms (Caenorhabditis elegans) has been shown to increase lifespan1,2. Such dysfunction triggers multiple signalling cascades, including a pathway called the mitochondrial unfolded protein response3 (UPRmt), which causes changes in gene expression that enable cells and organisms to cope with stress4. Writing in Cell, Tian et al.5 and Merkwirth et al.6 identify factors that relay mitochondrial dysfunction to the UPRmt.
Changes in gene expression are largely brought about by alterations in how tightly DNA is packaged around histone proteins in a structure called chromatin. Chromatin regulation is achieved partly through chemical decorations that modify histones, modulating this packaging. The current studies reveal that histone-modifying enzymes are integral players in the UPRmt stress response and lifespan extension in C. elegans.
By using genetic screens, Tian and colleagues found that the gene lin-65 is essential for the induction of UPRmt in response to mitochondrial stress. However, it should be noted that lifespan was extended even in the absence of lin-65, suggesting that mitochondrial stress also induces other pathways that promote longevity. The authors demonstrated that mitochondrial stress causes the LIN-65 protein to shuttle from the cytoplasm to the nucleus, reminiscent of the UPRmt regulator and transcription factor DVE-1, which also undergoes stress-induced nuclear migration7. Indeed, the researchers found that LIN-65 is required for the nuclear relocalization of DVE-1.
Tian et al. provided evidence to suggest that the addition of two methyl groups to aminoacid residue lysine 9 of histone H3 (a modification dubbed H3K9me2) in the cytoplasm by the MET2 methyltransferase enzyme triggered DVE-1 to shuttle to the nucleus. However, the possibility that non-histone targets of methylation are involved in this process cannot be ruled out, because the authors’ suggestion is based on methyltransferase inactivation, which could alter the methylation of many other proteins besides histones. The mechanism by which H3K9me2 promotes DVE-1 nuclear migration remains unknown.
The H3K9me2 modification is generally associated with tightly packaged chromatin. Indeed, Tian and colleagues observed that nuclei in worms exposed to mitochondrial stress consistently looked smaller and more dense than those in unstressed worms. DVE-1 localized to discrete spots away from DNA-dense regions, suggesting that this factor preferentially targets DNA regions that are less tightly packaged. This work supports a model whereby stress, through H3K9me2, induces changes in chromatin packaging that restrict the number of DNA regions to which DVE-1 can bind to regulate transcription. Thus, stress leads to specific changes in gene expression (Fig. 1).
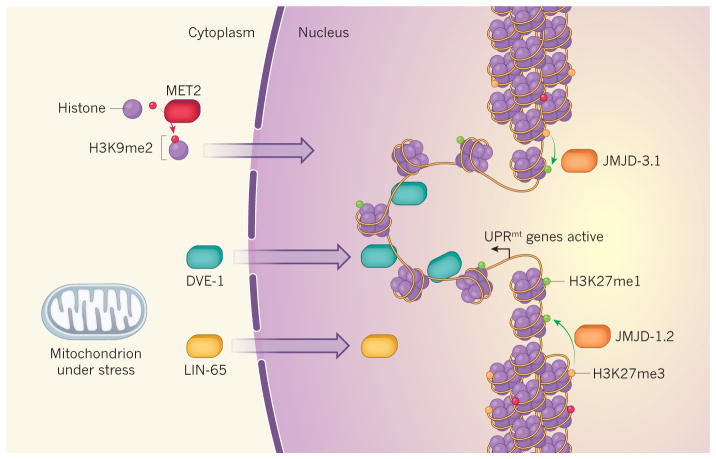
Figure 1. Coordinated responses to mitochondrial stress.
Gene expression can be inhibited by the tight packaging of DNA as chromatin around histone proteins that are decorated with methyl groups at specific amino-acid residues. Two such repressive modifications are dubbed H3K9me2 and H3K27me3. Two studies5,6 reveal that mitochondrial stress triggers the unpackaging and expression of genes involved in a stress response called the UPRmt. Tian et al.5 find that movement of the protein LIN-65 from the cytoplasm to the nucleus triggers nuclear migration of the transcription factor DVE-1. The enzyme MET2 mediates the addition of H3K9me2 modifications to histones in the cytoplasm; these histones move to the nucleus, restricting the unpackaged regions where DVE-1 can bind to promote transcription. Merkwirth et al.6 show that JMJD-1.2 and JMJD-3.1 enzymes are upregulated in response to mitochondrial stress and remove methyl groups from H3K27me3 to form the transcription-promoting modification H3K27me1 in the vicinity of UPRmt genes.
In a complementary study, Merkwirth et al. identified the genes jmjd-1.2 and jmjd-3.1 as crucial for the activation of the UPRmt pathway and extended lifespan in worms that had mitochondrial impairment. These genes encode enzymes that remove methyl groups from a histone modification dubbed H3K27me3, which is associated with tightly packaged chromatin. Mitochondrial impairment increases the expression of these two histone demethylases, which presumably leads to unpackaging of chromatin (Fig. 1). Overexpression of jmjd-1.2 and jmjd-3.1 in C. elegans recapitulates many of the effects of mild mitochondrial stress, including increased longevity and UPRmt induction, perhaps because increased levels of JMJD-1.2 and JMJD-3.1 repress the expression of many mitochondrial genes, thereby impairing mitochondrial function.
In both studies, the histone modifiers seemed to exert their effects predominantly on juvenile worms. It is therefore tempting to speculate that chromatin is most receptive to stress-induced reorganization in early life, and that these changes help to establish a stress response that persists into adulthood. Histone modifications can be stably maintained through cell divisions, even through generations, making them good candidate substrates for a ‘cellular memory’ of mitochondrial dysfunction. It will be interesting to determine whether H3K9me2 methyltransferases and H3K27me3 demethylases are required for maintenance of the mitochondrial-dysfunction-induced transcriptional program and activation of the UPRmt through to adulthood, even when the mitochondrial stress that triggered it is removed.
Because regulation of gene expression is already known to be central to the response to mitochondrial stress8, the participation of histone modifiers is not in itself unexpected. However, the current studies suggest that chromatin-modifying enzymes can be highly selective, eliciting programs of gene expression that are specific to a particular stress or pathway. Patterns of histone modifications can be extremely diverse, so it is not difficult to imagine how such exquisite specificity can arise. There are hundreds of histone modifications and many enzymes that have distinct and overlapping roles in depositing and removing them. DNA is wound around a nucleosome complex composed of four different histones, and each histone can be modified at many positions, so the number of possible combinations of histone modifications on each nucleosome is huge. Moreover, each nucleosome can have distinct modifications from its neighbours, building up an astronomically complex ‘histone code’.
Both groups found that the regulation of chromatin in response to mitochondrial stress is tissue specific. Tian et al. showed that LIN-65 activates the UPRmt pathway in the intestine, whereas Merkwirth et al. reported that JMJD-1.2 and JMJD-3.1 trigger the same response in neurons. An intriguing possibility is that distinct chromatin-regulating factors in different cells and tissues sense mitochondrial stress and respond in divergent yet coordinated ways that best enable organisms to counter the stress. It will be crucial to elaborate how stress signals can be coupled to chromatin regulation, and how cell-type-specific regulation can be coordinated. Perhaps mitochondrial impairment activates signalling cascades that regulate the expression, movements, modifications and activities of distinct histone modifiers in specific cells. Mitochondrial impairment could also result in altered levels of metabolic intermediates, many of which act as essential co-factors of histone-modifying enzymes.
The idea that a stress signal can engage specific histone modifiers to elicit a persistent response will no doubt extend to diverse stressors. Indeed, JMJD-3.1 has been implicated9 in the dampening of a stress response called the heat-shock response from early adulthood through to ageing in C. elegans. Additionally, by-products of cellular metabolism called free radicals modulate longevity in yeast through a histone-modifying enzyme10. We can look forward to many more exciting insights into the role of chromatin regulation in stress responses.
Contributor Information
Siu Sylvia Lee, Department of Molecular Biology and Genetics, Cornell University, Ithaca, New York 14850, USA.
Jessica K. Tyler, Department of Pathology and Laboratory Medicine, Weill Cornell Medicine, New York, New York 10065, USA
References
- 1.Dillin A, Crawford DK, Kenyon C. Science. 2002;298:830–834. doi: 10.1126/science.1074240. [DOI] [PubMed] [Google Scholar]
- 2.Rea SL, Ventura N, Johnson TE. PLoS Biol. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haynes CM, Ron D. J Cell Sci. 2010;123:3849–3855. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- 4.Durieux J, Wolff S, Dillin A. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian Y, et al. Cell. 2016;165:1197–1208. doi: 10.1016/j.cell.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merkwirth C, et al. Cell. 2016;165:1209–1223. doi: 10.1016/j.cell.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. Dev Cell. 2007;13:467–480. doi: 10.1016/j.devcel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 8.Chang HW, Shtessel L, Lee SS. Free Radic Biol Med. 2015;78:168–178. doi: 10.1016/j.freeradbiomed.2014.10.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Labbadia J, Morimoto RI. Mol Cell. 2015;59:639–650. doi: 10.1016/j.molcel.2015.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schroeder EA, Raimundo N, Shadel GS. Cell Metab. 2013;17:954–964. doi: 10.1016/j.cmet.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]