Figure 6. Expected output of code provided in the worked example.
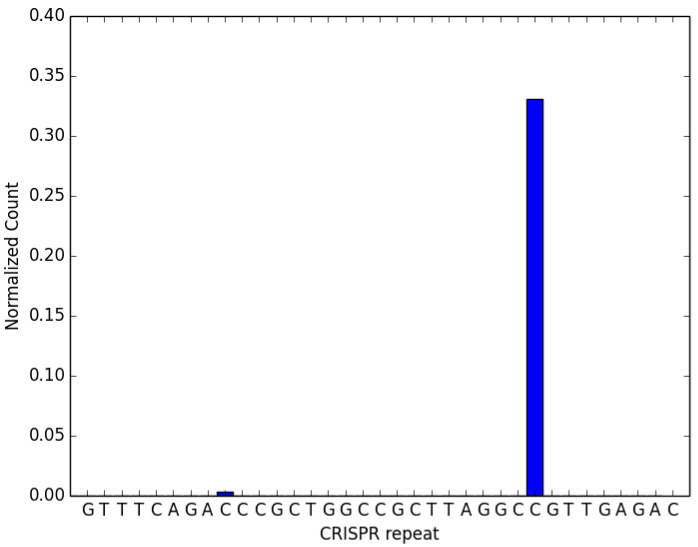
. Processed crRNA levels assayed by high throughput small RNA sequencing. This dataset has been artificially supplemented with sequences matching expected CRISPR-derived RNAs. The CRISPR repeat sequence from the 1st line of the parameters file crRNAfigureMaker_params.txt is on the X-axis. The height of the bar at each base along the X-axis represents the relative proportion of crRNAs with 3’ ends at that base, normalized to the levels of the reference RNA (isoleucine tRNA; consistently the most abundant species encountered in our M. mediterranea datasets). The presence of a distinct 3’ end sequence in the population of CRISPR repeat containing RNAs indicates site-specific cleavage and processing of pre-crRNA.