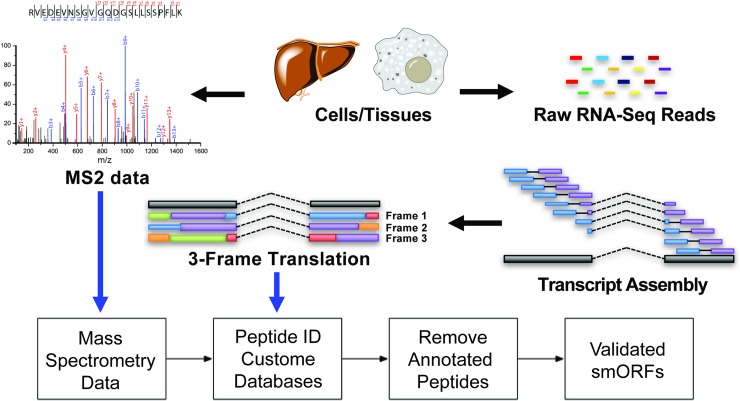
Fig 1. Proteogenomics microprotein discovery pipeline.
High quality RNA-Seq data is assembled and three-frame translated to create an in silico custom proteomics database. The database is then used to search MS2 spectra to obtain peptide candidates. Short un-annotated peptides with a high quality MS2 spectra are manually curated to produce a list of novel confident microproteins. Since peptide discovery depends on the assembled transcriptome, we propose to optimize transcriptome assembly for peptide discovery.