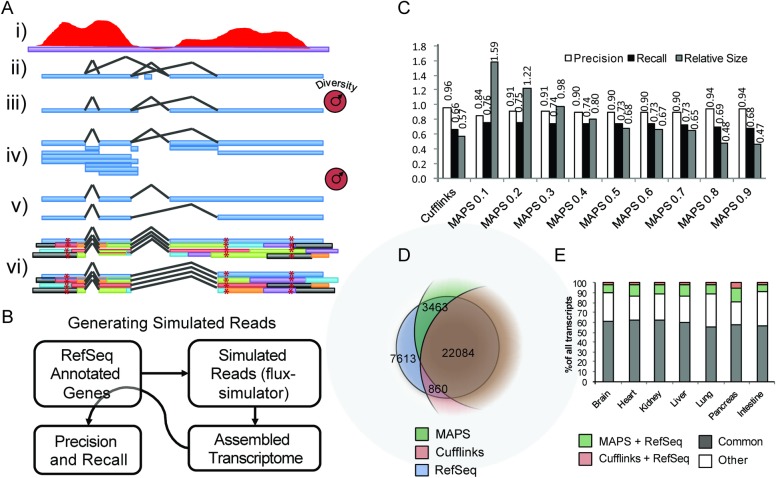
Fig 2. Building and testing a tunable ORF assembler.
A) Overview of MAPS, A novel tunable ORF assembler. High-depth paired-end reads (i) are used to identify all possible supported contigs and junctions (ii), which are then filtered (iii) and used to generate all possible supported exons (iv). Transcripts are then assembled using greedy heuristics and filtered based on read support (v). The resulting transcripts are then in silico translated for all frames to produce a set of supported ORFs using the consensus read sequence and the annotated genomes to further extend the ORFs (vi). B) Workflow for determining precision and recall of the assembled transcripts using realistic simulated RNA-Seq reads generated from annotated RefSeq genes. C) Simulated reads were assembled using a generalized assembler, Cufflinks, and MAPS with varying stringency (diversity) settings, and the precision, recall, and relative size of the resulting transcriptomes are shown. D) Plot showing the relative fractions of RefSeq transcripts assembled with MAPS (Stringency = 0.3) and Cufflinks using the simulated reads workflow in B). E) ENCODE paired end mouse RNA-Seq datasets for varying tissues were assembled using MAPS and Cufflinks and fractions of shared transcripts are shown. Colors represent transcripts overlapping between just MAPS assembly and RefSeq annotated genes (green), transcripts overlapping between just Cufflinks and RefSeq annotated genes (pink), transcripts overlapping between RefSeq, MAPS, and Cufflinks (gray), and all other transcripts that are assembled but not annotated in RefSeq (white).