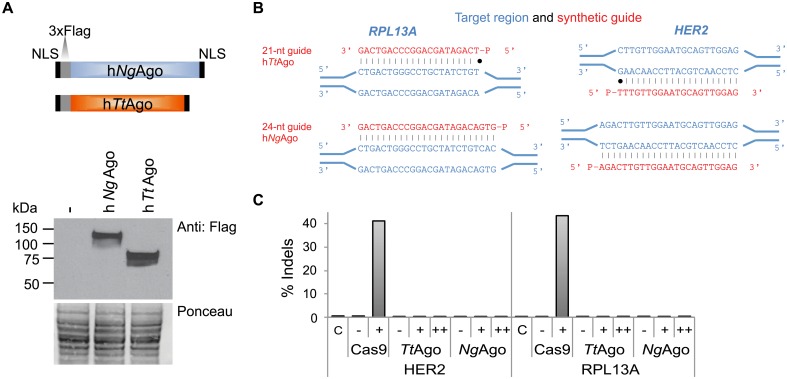
Fig 2. hTtAgo and hNgAgo do not cleave genomic target sites.
A. Schematic of human codon-optimized TtAgo and NgAgo proteins containing two nuclear localization signals (NLSs) and a 3xFlag epitope tag. Western blot analysis of hTtAgo and hNgAgo proteins in HEK293 cells using an antibody against the 3xFlag tag. Untransfected cells serve as a negative control (-). Ponceau staining was used as a loading control. B. Diagram illustrating RPL13A and HER2 guide DNAs and genomic target site. Genomic target sites are indicated in blue and complementary ssDNA guides are indicated in red. C. Amplicon sequencing was carried out on HEK293 cells co-transfected with hTtAgo or hNgAgo expression plasmids with (+) or without (-) gDNAs to RPL13A and HER2. To increase the amount of gDNAs, cells were re-transfected with gDNAs 24 hours after the initial transfection (++). CRISPRESSO analysis confirms that hTtAgo and hNgAgo did not cause insertions or deletions (indels) under any of these conditions (S5 Table). As a control, HEK293 cells were co-transfected with Cas9 nuclease and gRNA expression plasmids targeting RPL13A and HER2. RNA-guided Cas9 displayed target site cleavage at the genomic RPL13A and HER2 target sites. The percentage of sequence reads containing indels relative to the total number of sequence reads is plotted on the y-axis.