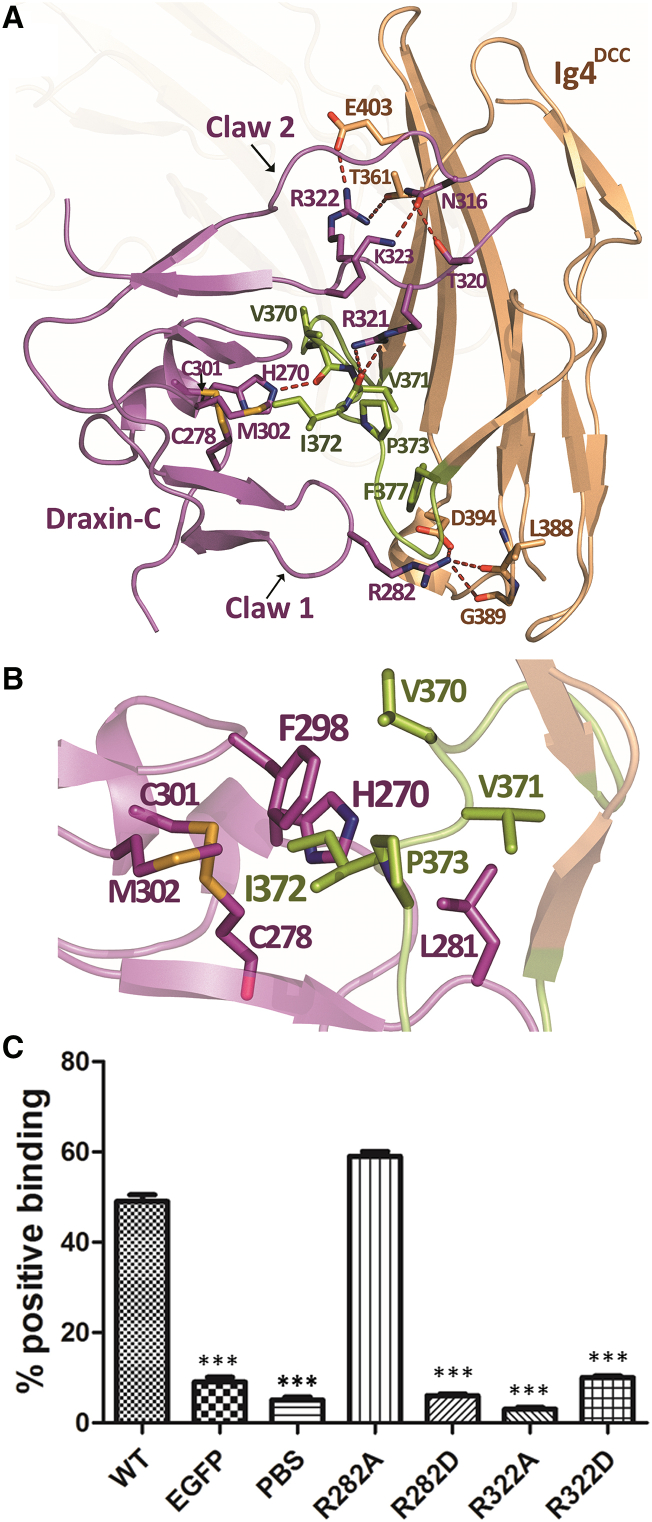
Figure 2.
Characterization of Draxin/DCC Binding
(A) Overview of the interactions between rDraxin-C and the Ig4 domain of rDCCIg1Ig4. Residues mediating important interactions between the two proteins are shown as sticks and labeled. Salt bridge and hydrogen bond interactions are shown as dashed lines. The CD loop on the Ig4 domain of DCC is colored in green.
(B) Detailed view of the hydrophobic hotspot of the rDraxin-C/r DCCIg1Ig4 complex. Residue Ile372 from the CD loop (colored in lemon) of rDCC-Ig4 stacks between His270 and Phe298 of rDraxin-C to form the core of the hydrophobic cluster. The core is surrounded by the hydrophobic side chains of Val370, Val371, and Pro372 from the CD loop of DCCIg1Ig4 and Cys278, Leu281, Cys301, and Met302 from rDraxin-C. The view is rotated 45 degrees along the vertical axis with regard to (A).
(C) Cell-binding assays for full-length wild-type (WT) rDCC showing the percentage of DCC presenting HEK293Tcells that bind rDraxin WT and mutants Arg282Ala, Arg282Asp, Arg322Ala, and Arg322Asp. As a control, eGFP and PBS were used. Data represent mean ± SE (n = 100 for each group). One-way ANOVA, followed by a post hoc Scheffé’s test, was performed. ∗∗p < 0.001 compared with WT.