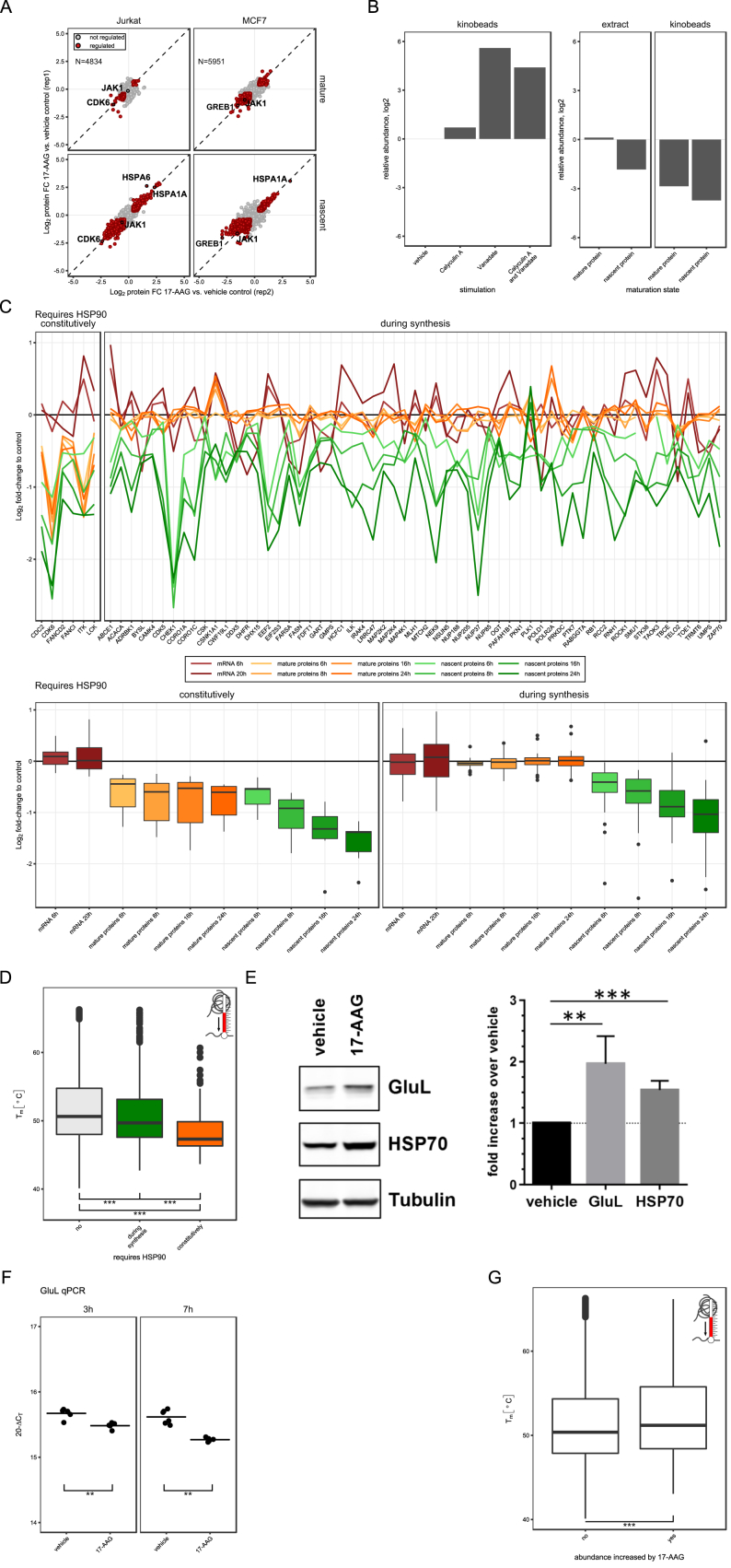
Figure S5.
Validation of Effects Observed upon HSP90 Inhibition, Related to Figure 5
(A) Scatterplots showing protein fold changes (FC) observed in mature (upper panel) and nascent (lower panel) forms of proteins in Jurkat cells (left panel) and MCF-7 cells (right panel) treated for 24 h with 17-AAG relative to vehicle treated cells. Red closed circles indicate statistically significant regulation (p < 0.05) and the dashed diagonal indicates the equality line. N indicates the number of proteins robustly quantified in both replicates.
(B) Protein fold change (FC) of ZAP70, following kinobeads affinity enrichment in cell extracts generated from Jurkat cells preincubated for 30 min with the phosphatase inhibitors calyculin A (0.05 μM) and orthovanadate (30 μM), individually or in combination (left panel) relative to vehicle treated cells. Protein fold change of mature and nascent ZAP70 after 24 h incubation with 17-AAG (central panel) and subsequent kinobeads affinity enrichment (right panel).
(C) Comparison of relative mRNA levels as reported in Fierro-Monti et al. PloS ONE 2013 with the mPDP dataset of mature and nascent relative protein levels generated in Jurkat cells after treatment with 17-AAG at different time points for a subset of genes referred to in both datasets (upper panel). Boxplot representation showing log2 FC for mature, nascent protein and mRNAs (same subset displayed in the upper panel) for different 17-AAG treatments (lower panel).
(D) Boxplots of proteins thermal stabilities in Jurkat cells (displayed as Tm in degrees Celsius) grouped according to HSP90 dependence, synthesis or constitutive (∗ p ≤ 0.05, ∗∗ p ≤ 0.01 and ∗∗∗ p ≤ 0.001). Melting points (Tm) were determined by thermal proteome profiling.
(E) Representative protein immunoblots of GLUL, HSP70 and Tubulin in Jurkat cells treated for 6 h with 10 μM 17-AAG. Bar chart: Quantification of band intensity from 3 independent experiments (mean with SD) displayed as fold increase normalized to control vehicle (∗ p ≤ 0.05, ∗∗ p ≤ 0.01 and ∗∗∗ p ≤ 0.001).
(F) qPCR of GLUL mRNA levels by in Jurkat cells after treatment with 17-AAG (10 μM) for 3 and 7 hr. Data are displayed as 20-ΔCT (cycle times for GLUL amplification normalized to housekeeping gene RPL7) of two independent experiments performed in triplicate (∗ p ≤ 0.05, ∗∗ p ≤ 0.01 and ∗∗∗ p ≤ 0.001).
(G) Boxplots of thermal stabilities (Tm) of proteins with significantly increased abundance by upon 17-AAG treatment in Jurkat cells (right) compared to proteins that do not change (left) at any measured time point (6-24 h). Only those proteins for which Tms could be determined are included (∗∗∗ p ≤ 0.001).