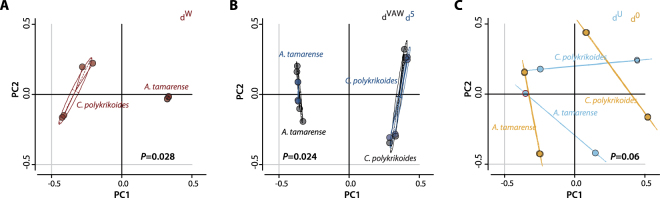
Figure 2.
Comparison of the phylogenetical differences between samples. The generalized UniFrac PCoA plots grouped with the associated microalgae as the discriminating factor. The PCoA plots are based on (A) weighted UniFrac distance (dW), (B) moderately-weighted UniFrac distance (dVAW), and (C) unweighted UniFrac distance (dU). d0.5 and d0 indicate the generalized UniFrac distance of d(α), where α controls the contribution of high-abundance branches ranging from 0 to 1. The perMANOVA p-value represents the statistical significance between microbial communities identified from A. tamarense and C. polykrikoides, regardless of the sequencing method.