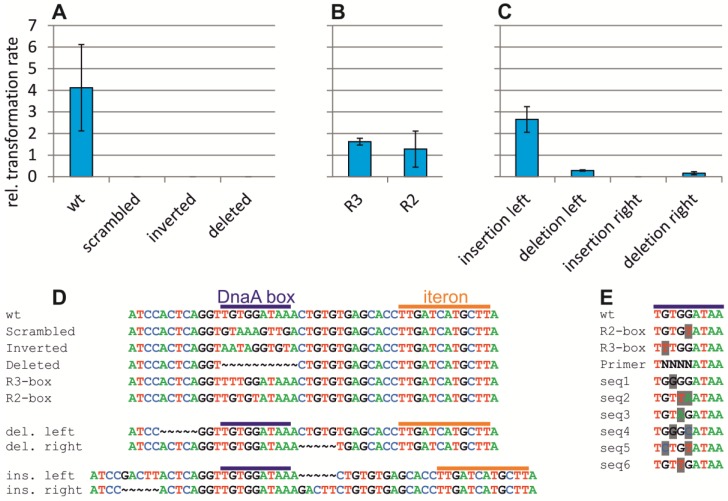
Figure 5.
Mutation-based analysis of the DnaA box within ori2. (A–C) Ori2-based minichromosomes with different mutations at the DnaA box were tested for their ability to replicate by transformation into E. coli (XL1-Blue) or DH5αλpir. The latter was used as a control because all replicons carry an oriR6K which allows replication in strains encoding the initiator Pir. Values are ratios between respective colony numbers of three biological replicates with the indicated standard deviations. Relevant sequences are shown in (D) for DnaA boxes wt (pMA87), scrambled (pMA108), inverted (pMA109), deleted (pMA110), a weak DnaA box R3 (pMA111), a medium-strength DnaA box R2 (pMA112), a 5 bp insertion to the left of the DnaA box (pMA115) or the right (pMA114) or a 5 bp deletion to the left (pMA113) or right (pMA116) as indicated. (E) Sequences found by transformation of an assembled ori2-minichromosome with a mix of sequence combinations at positions 2–5 as indicated by “N” in the Primer sequence. DnaA-box sequences from oriC in E. coli are shown for comparison (wt, R2, R3). Six sequences found in the screen are shown with the nucleotides differing from the consensus DnaA box shaded in grey.