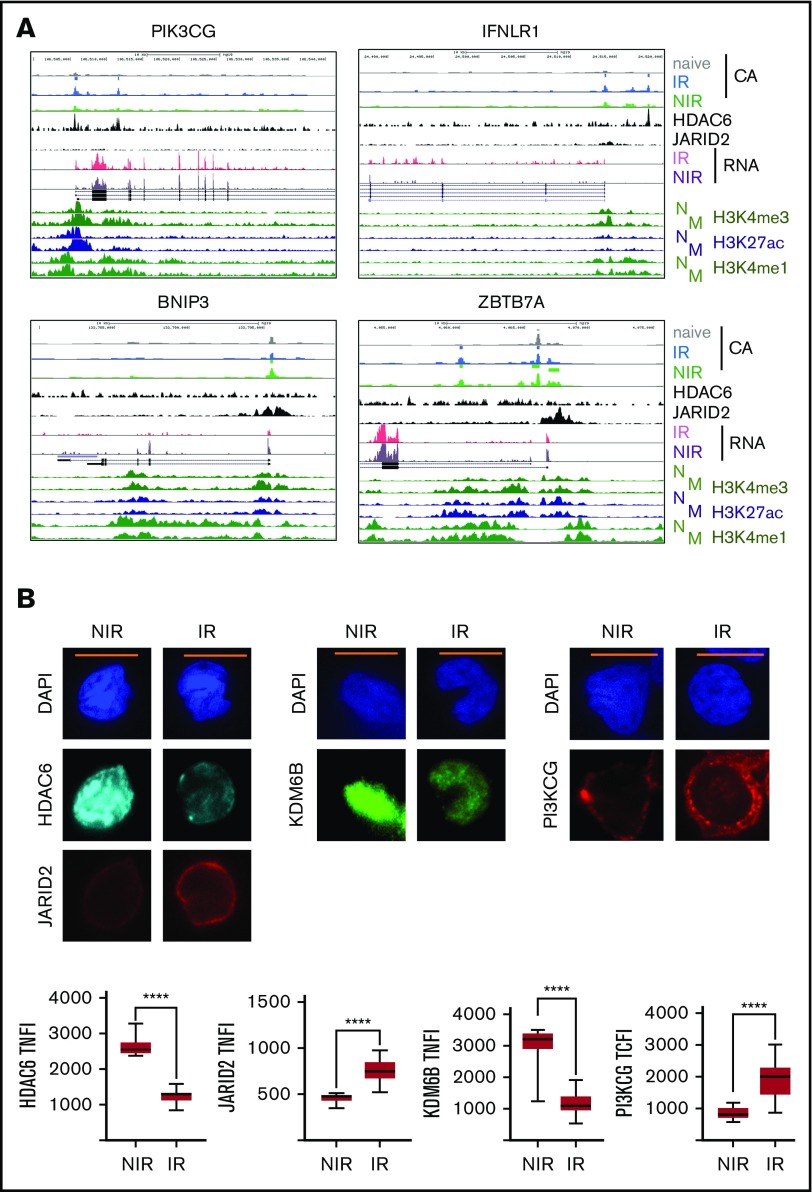
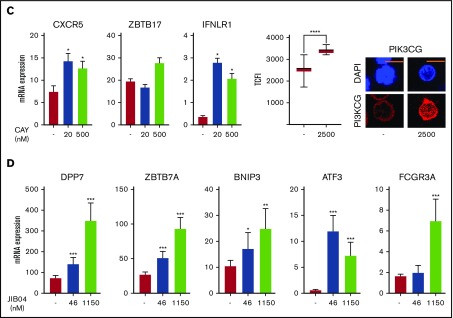
Figure 5.
Regulation of gene expression in IR and NIR lymphocytes by chromatin modulators. (A) Chromatin accessibility (CA; FAIRE-seq), mRNA levels (RNA-seq), and HDAC6, JARID2, and histone modifications (publicly available ChIP-seq) near PIK3CG, IFNLR1, BNIP3, and ZBTB7A. Histone modifications are for naive (N) and memory (M) CD8+ lymphocytes (REMC/Broad donor 100). HDAC6 binding in activated T lymphocytes and JARID2 binding in iPSCs. (B) Levels and location of HDAC6, JARID2, KDM6B, and PI3KCG proteins in IR and NIR cells. (C) The effect of pretreatment with the HDAC6 inhibitor CAY1063 on gene expression levels and PIK3CG protein expression in CD8+ lymphocytes following 2-hour phorbol 12-myristate 13-acetate/ionomycin stimulation. (D) The effect of pretreatment of unstimulated CD8+ lymphocytes with the JARID inhibitor JIB04 on gene expression levels. (B-C) Total nuclear fluorescence intensity (TNFI) or total cytoplasmic fluorescence intensity (TCFI) was measured in a minimum of n = 20 cells for 2 different donors. DAPI (4′,6-diamidino-2-phenylindole) shows the nucleus. ****P < .0001. Scale bar represents 5 μm. (C-D) Expression values normalized to PPIA. Error bars show standard error of the mean of triplicate polymerase chain reactions from 1 donor and are representative of 2 different donors. ***P < .001, **P < .01, and *P < .05 compared with control.