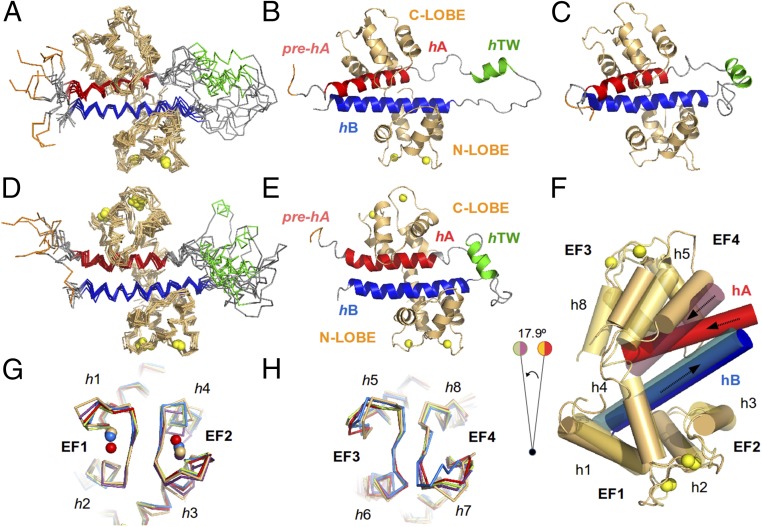
Fig. 2.
Structure of the CaM/Kv7.2-hAB complex at different calcification states. An overlay of the 10 lowest energy conformers representing the 3D solution structure of the intCaM/Kv7.2-hAB (A) or holoCaM/Kv7.2-hAB (D) complex is shown. A ribbon representation of the intCaM/Kv7.2-hAB (B) or holoCaM/Kv7.2-hAB (E) complex shows the structural elements: CaM protein (orange); the prehelix A(pre-hA), which connects segment 6 of the pore with the rest of the intracellular C terminus of the Kv7.2 channel (purple); helix hA (red); helix hTW (green); and helix hB (blue). Ca2+ ions are represented as spheres. The same color code is used for the ribbon representations of the apoCaM/Kv7.2-hAB complex. (C) Ribbon representation of the structural model of the apoCaM/Kv7.2-hAB complex. (F) Alignment of intCaM/Kv7.2-hAB and holoCaM/Kv7.2-hAB by the center of mass of the N-lobe of CaM displays the rotation (17.9°) of a segment of the complex upon Ca2+ binding to the C-lobe of CaM. The involved structural elements are h5 and h8 in CaM and hA in Kv7.2-hAB. (G) Alignment of the EF-hand motifs of the N-lobe of intCaM/Kv7.2-hAB (orange) with the equivalent motif in other reported (Ca2+-loaded) CaM structures: 4RJD (purple), 4UMO (blue), 4GOW (red), and 5J03 (green), with rmsd values of 1.12 Å, 1.22 Å, 1.26 Å, and 1.39 Å, respectively. (H) Alignment of the EF-hand motifs of the C-lobe of intCaM/Kv7.2-hAB (orange) with the equivalent motif in other reported (apo) CaM structures: 4V0C (blue), 4E50 (red), 4JQ0 (purple), and 5J03 (green), with rmsd values of 0.93 Å, 0.94 Å, 1.18 Å, and 1.25 Å, respectively.