Figure 3.
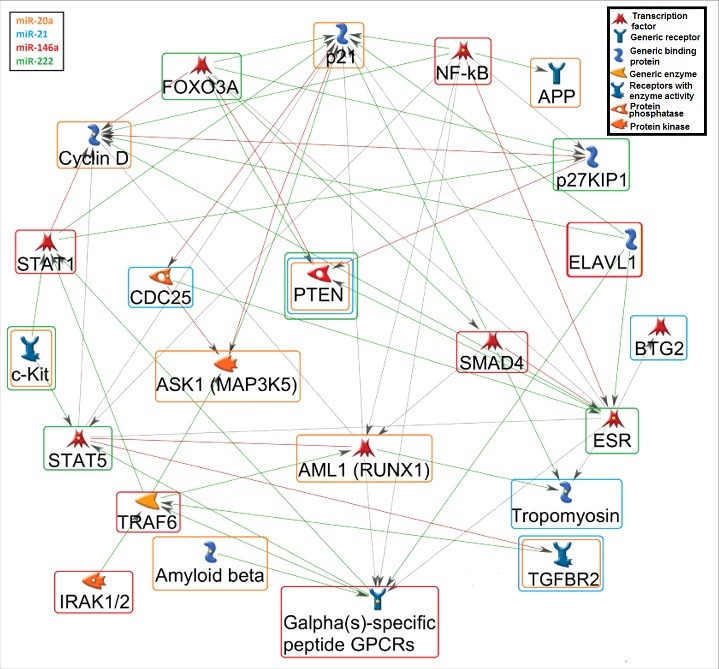
Gene network among the putative miRNA targets. A gene network (MetaCore™) was generated for the potential connections of at least two miRNA-targets. The orange rounded rectangle corresponds to miR-20a, blue to miR-21, red to miR-146a, and green to miR-222 targets. The green arrows show activation, the red arrows indicate inhibition, and the grey arrows are unspecified connections. Details for the genes shown in the figure: AML1 (RUNX1): Runt-related transcription factor 1; APP: Amyloid beta (A4) precursor protein; ASK1 (MAP3K5): Mitogen-Activated Protein Kinase Kinase Kinase 5; BTG2: BTG family, member 2; CCND: Cyclin D; CDC25: Cell Division Cycle 25; CDKN1A (p21): Cyclin-Dependent Kinase Inhibitor 1A; CDKN1B (p21KIP1): Cyclin-dependent kinase inhibitor 1A/B; ELAVL1: ELAV Like RNA Binding Protein 1; ESR: Estrogen receptor; FOXO3A Forkhead box O 3A; GPCRs (CXCR4): Chemokine receptor 4 (G Protein-Coupled Receptors); IRAK1/2: Interleukin-1 Receptor-Associated Kinase 1/2; c-KIT: v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog; NFKB: Nuclear factor of kappa light polypeptide gene enhancer in B-cells; PTEN: Phosphatase and tensin homolog; SMAD4: SMAD family member 4; STAT1/5: Signal Transducer And Activator Of Transcription 1/5; TGFBR2: Transforming growth factor, beta receptor II; TPM1: Tropomyosin; TRAF6: TNF receptor-associated factor 6.
