Figure 2.
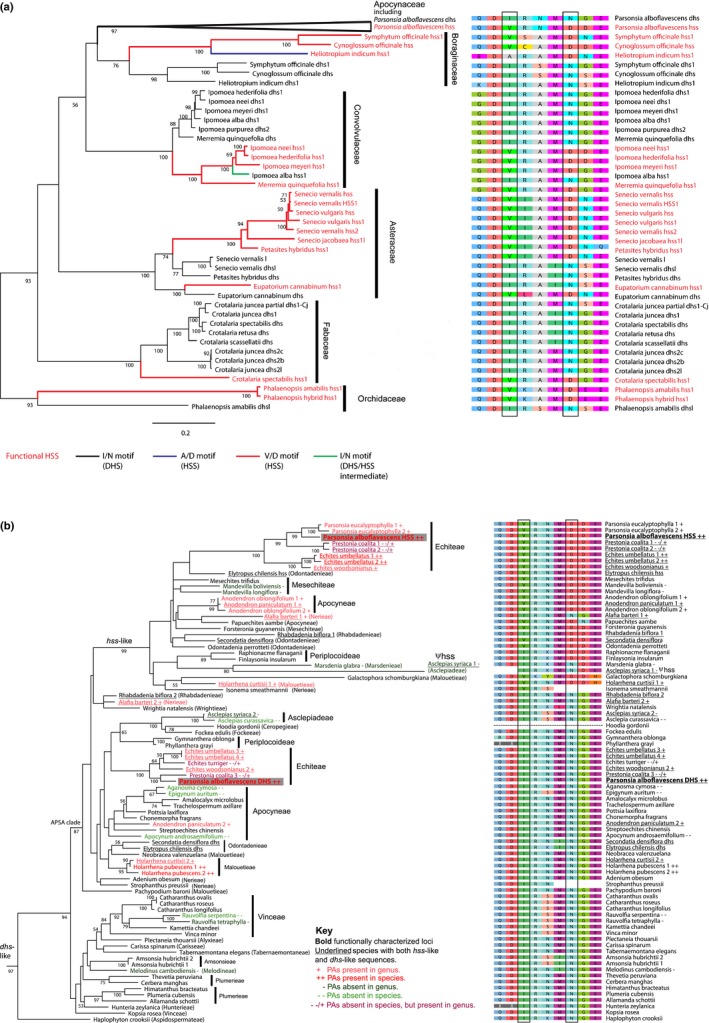
Maximum‐likelihood gene tree with bootstrap support of (a) all pyrrolizidine alkaloid (PA)‐producing genera with functionally characterized homospermidine synthase (HSS) loci and (b) candidate deoxyhypusine synthase (dhs)/hss loci from Apocynaceae. The evolution of the amino acid motif at positions 305 and 308 (numbered based on our alignment Supporting Information Notes S1) with maximum marginal likelihood is shown and the alignment is illustrated. (a) Functionally characterized HSS and DHS loci from across angiosperms. (b) Apocynaceae hss/dhs‐like loci. Red text, names of functionally characterized HSS sequences; black lines, isoleucine/asparagine (I/N) motif; red lines, valine/aspartic acid (V/D) motif; purple lines, alanine/aspartic acid (A/D) motif; blue lines, isoleucine/aspartic acid (I/D) motif; green lines, secondarily derived isoleucine/asparagine (I/N) motif. Underlined names, Apocynaceae species with both putative dhs and putative hss; +, PAs present in genus, unknown in species; ++, PAs present in species; ‐, PAs absent in genus, unknown in species; ‐ ‐, PAs absent in species; ‐ ‐/+, PAs absent in species but present in genus.
