Figure 4.
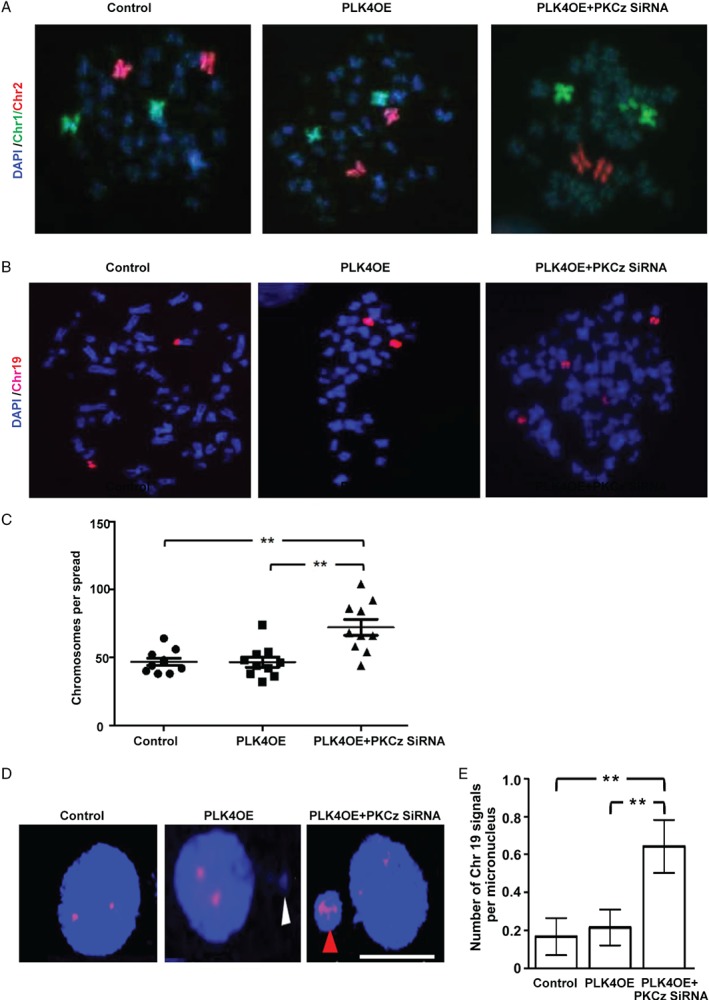
Effects of PKCz on chromosome segregation in cells with extra centrosomes. (A) Chromosome (Chr) 1 (green) and 2 (red) signals in control Caco‐2 transfected with empty vector only versus PLK4OE versus PLK4OE + PKCz siRNA‐transfected Caco‐2 cells. Chromosome fluorophores were counterstained against the DAPI DNA stain (blue). Note 2 × Chr1 and 2 × Chr2 signals in control and PLK4OE cells but 3 × Chr1 signals in Caco‐2 PLK4OE + PKCz siRNA‐transfected cells. We analysed 20 spreads per experimental condition. We found >2 Chr1 and/or >2 Chr2 signals in 1/20 control Caco‐2 and Caco‐2 PLK4OE spreads each. However, 4/20 Caco‐2 PLK4OE‐PKCz siRNA spreads showed >2 Chr1 and/or >2 Chr2 signals on FISH assay. (B) Chromosome 19 (red) signals in control Caco‐2 versus PLK4OE versus PLK4OE + PKCz siRNA‐transfected Caco‐2 cells (n = 30 cells per spread). We found >2 Chr19 signals in 9/30 control Caco‐2, 10/30 PLK4OE, and 14/30 Caco‐2 PLK4OE + PKCz siRNA‐transfected Caco‐2 cells. (C) Total chromosome number per spread in control Caco‐2 versus PLK4OE versus PLK4OE + PKCz siRNA‐transfected Caco‐2 cells, **p < 0.01; ANOVA; Tukey's post hoc test; control Caco‐2 versus PLK4OE = NS (n = 10 spreads per experimental condition). (D) Chromosome 19 (red) signals and micronuclei in control Caco‐2 versus PLK4OE versus PLK4OE + PKCz siRNA‐transfected Caco‐2 cells. Micronuclei lacking or containing a Chr19 signal are indicated by white or red arrowheads, respectively. (E) Summary of Chr19 signals per micronucleus in control Caco‐2 versus PLK4OE versus PLK4OE + PKCz siRNA‐transfected Caco‐2 cells, **p = 0.011; ANOVA with Tukey's post hoc test (Caco‐2 control versus Caco‐2 PLK4OE = NS). Scale bar = 20 μm.
