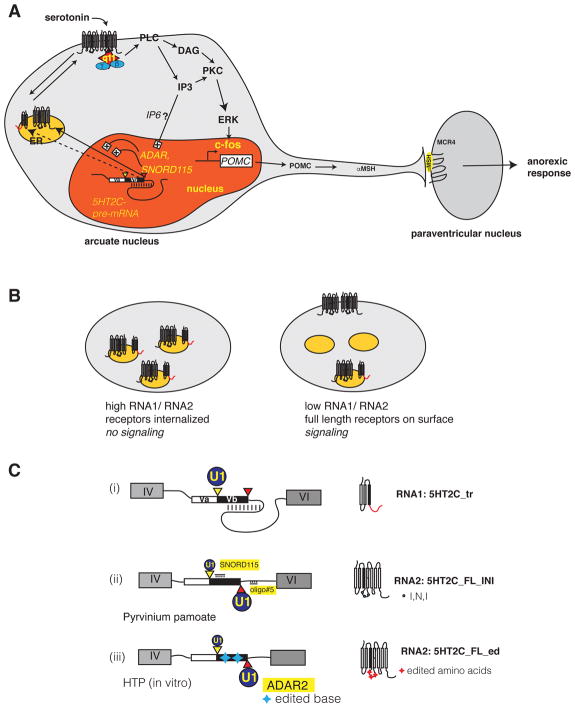
Figure 3. Biological function of 5HT2C isoforms.
A. Signaling of 5HT2C in POMC neurons of the arcuate nucleus
The regulated dsRNA structure shown in Figure 1C is indicated schematically in the nucleus (red). Yellow and red triangles indicate the proximal and distal splice sites. ADAR and SNORD115 promote the formation of the full-length receptor through inclusion of exon Vb. The full-length receptor shuttles to the plasma membrane where it is active as a dimer. A heterotrimeric G protein containing Gq11 (red star, blue circles) binds to the second intracellular loop. The binding affinity of the heterotrimeric G protein is regulated by the editing of three amino acids in the second intracellular loop (Figure 2B, E). Gq11 activates PLC, leading to the formation of DAG and IP3, which activates c-fos via the ERK pathway. C-fos turns on the POMC promoter, and the resulting POMC is processed into alpha MSH, which signals to neurons in the paraventricular nucleus generating a satiety signal.
ER: endoplasmic reticulum, q11: Gq11 alpha subunit, PLC: phospholipase C, IP3: inositol trisphosphate, PKC: protein kinase C, DAG: diacylglycerol, ERK: extracellular regulated kinase, POMC: pro opiomelanocortin, MSH: melanocyte stimulating hormone; MCR4: melanocortin receptor 4.
B. Regulation of 5HT2C signaling through heterdimerization of the splicing isoforms
The truncated 5HT2C receptor is localized in intracellular membranes, likely endoplasmic reticulum (orange circle). It heterodimerizes with full-length receptors likely through interaction of the first transmembrane domains. Thus a high ratio of RNA1 to RNA2 leads to an internalization of full-length receptors, which diminishes signaling. In contrast, a low RNA1/RNA2 ratio leads to full-length receptors on the surface and signaling.
C. The 5HT2C pre-mRNA as a therapeutic target
(i) The default splice site selection is usage of the proximal splice site (yellow triangle) due to the secondary structure blocking the distal site (red triangle). U1: U1 snRNP, the size of the circle indicates the binding strength
(ii) Oligo#5 binds to the intronic part of the dsRNA structure, activating the distal splice site, similar to SNORD115 that binds to the exonic part.
(iii) ADAR action also decreases the stability of the dsRNA structure, leading to distal splice site activation. Helix-threading peptides block ADAR2 activity in cell free assays, showing that this process can also be modified in principle (Schirle et al. 2010).