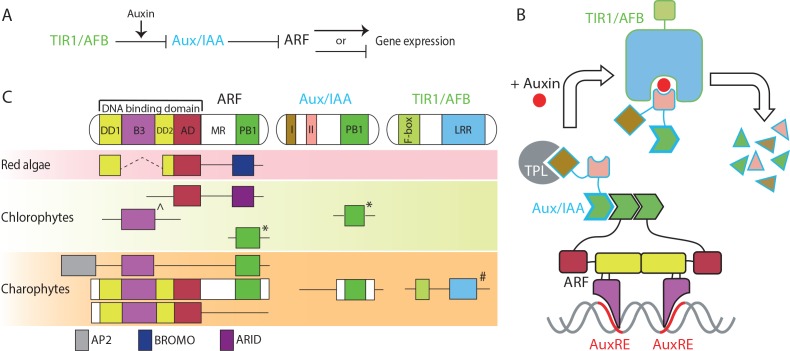
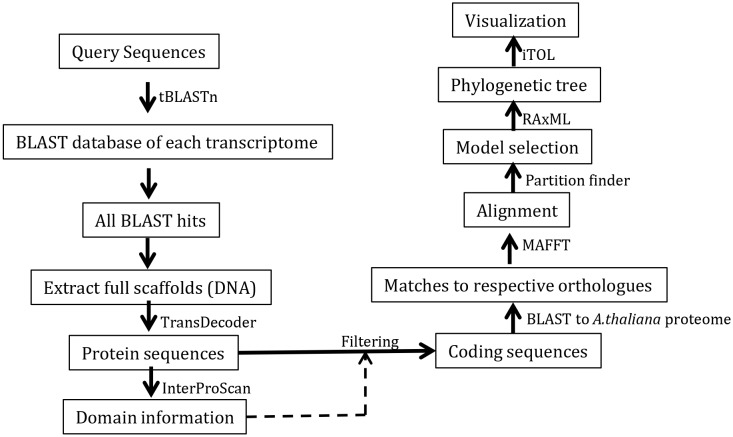
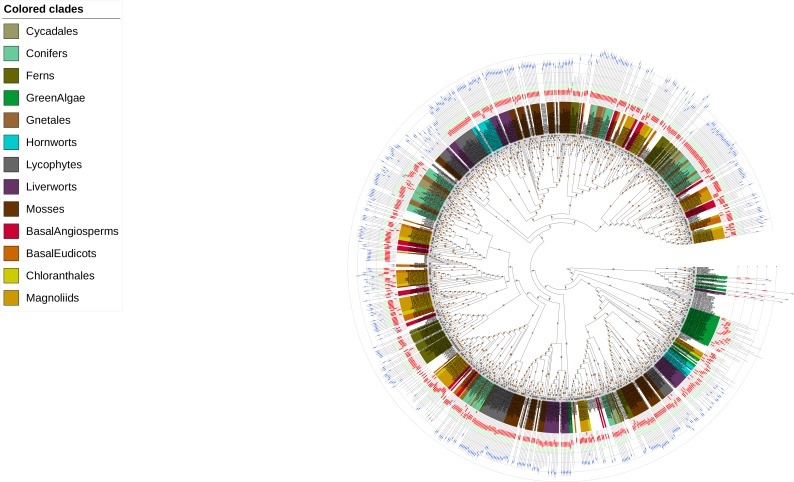
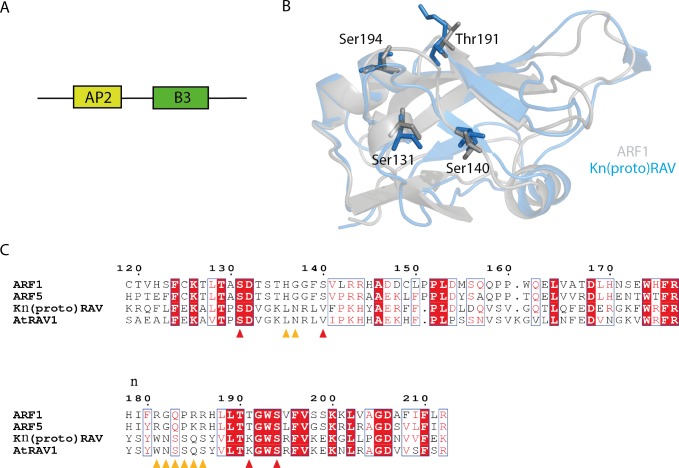
Figure 1. Proteins in nuclear auxin pathway; mechanism and origin of the domains.
(A, B) Scheme of NAP in land plants. In the absence of auxin, Aux/IAA inhibit ARF via their PB1 domains, and by recruiting the TPL co-repressor. Auxin stabilizes the interaction between Aux/IAA and TIR1/AFB, followed by proteasome-mediated degradation of Aux/IAA. (C) Domain structure of NAP components in land plants and presence of each domain in algae, as recovered in transcriptomes. (Sub-)domains are indicated by colors, that match those in (B). ^: basal to all B3-type transcription factors in land plants, *: difficult to assign to ARF or Aux/IAA family; #: forming basal clade to both TIR1/AFB and COI1 in land plants.