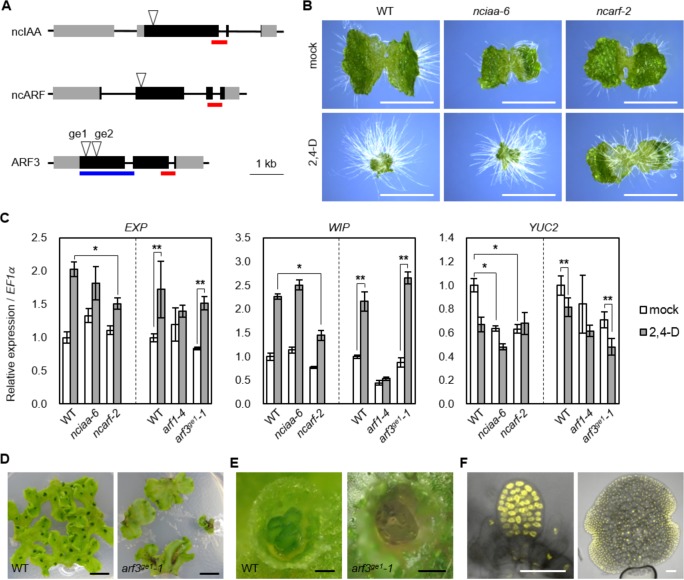
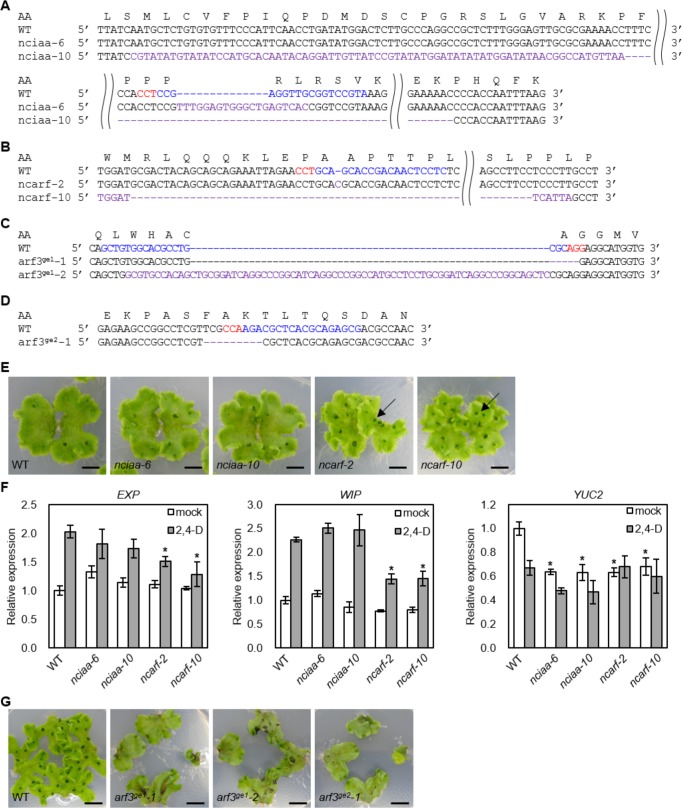
Figure 7. Genetic analysis of ancient components in Marchantia polymorpha.
(A) Diagrams of gene structure and CRISPR/Cas9-mediated mutation in ncIAA, ncARF and ARF3 loci. Arrowheads indicate sgRNAs target sites. Gray and black boxes indicate UTR and CDS, respectively. Red and blue bars indicate the region coding PB1 and DBD. (B) 10-day-old gemmalings grown without or with 3 μM 2,4-D. Scale bars: 5 mm. (C) Expression analysis of auxin-responsive genes in WT, nciaa, ncarf, and arf3 mutants by qPCR. 10-day-old gemmalings (nciaa and ncarf) or regenerating thalli (arf1 and arf3) were treated with 10 μM 2,4-D for 1 hr. Each bar indicates average ±SD (biological replicates = 3). Asterisks indicate significant differences. *: p<0.01 (Tukey test), **: p<0.05 (t-test). (D, E) Thallus tips grown for 2 weeks (D) and gemma cups (E) of WT and arf3ge1-1 mutant. arf3ge1-1 showed growth retardation and no mature gemmae, similar to the other alleles. (F) Expression analysis of proARF3:ARF3-Citrine in arf3ge2-1 background. Left and right panel show developing and mature gemmae, respectively. Scale bars: 5 mm in (B and D), 0.5 mm in (E), 50 μm in (F).