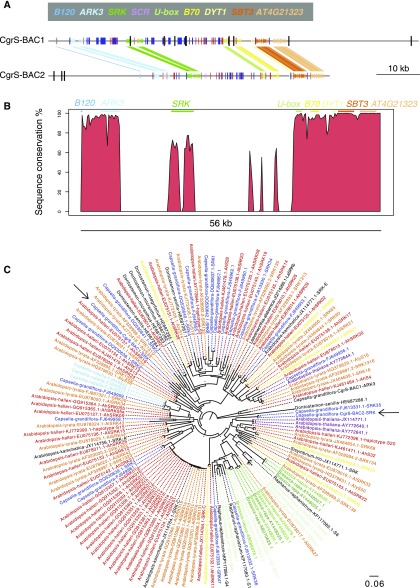
Figure 1.
A S-locus sequence assemblies with two measures of indel errors indicated in black bars. Inference of indel errors are based on comparison of two independent SMRT-sequencing runs and assemblies of CgrS-BAC1 (upper) and alignment of Illumina short reads to assembly of CgrS-BAC2 (lower). Annotation of exons are shown as colored arrows, simple repeat sequences in red, and blue-boxes indicate positions of transposable elements. The genes flanking the S-locus are ARK3 (light blue) and U-box (light green). SCR was only annotated in CgrS-BAC2. B S-locus sequence conservation between the two Capsella S-locus BACs, created by aligning the S-locus regions with LASTZ and comparing sequence homology (in % between 0 and 100) using a fixed window size of 250 bp. Sequence similarity between CgrS-BAC1 and CgrS-BAC2 drops steeply at the borders of the S-locus, corresponding to the genes ARK3 and U-box, respectively, although some sequence similarity is also found at SRK. C ML phylogeny of all alignable SRK alleles (exon 1) above 500 bp from GenBank. Bootstrap support over 70% is represented with an asterisk (*). Our newly identified sequences, indicated with arrows, are found broadly distributed across the phylogeny.