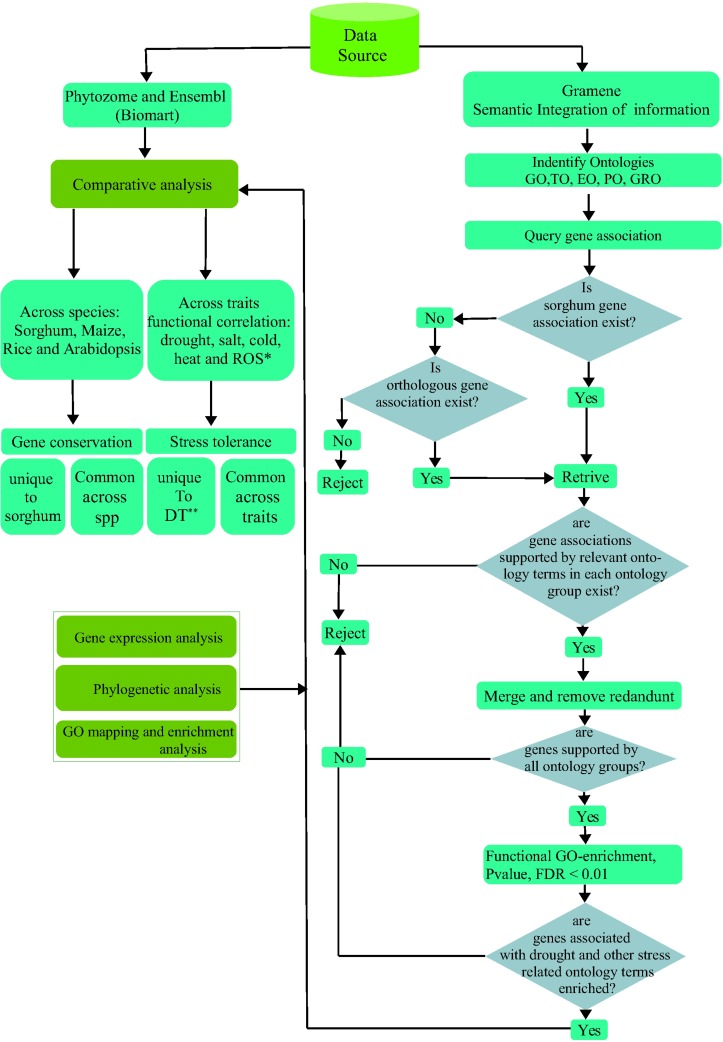
Fig 1. Work-flow for gene-phenotype association across-species and stresses.
This figure demonstrates the work flow for the gene-phenotype association in sorghum stress tolerance across species by comparing sorghum drought responsive genes with orthologs in maize, rice and Arabidopsis and across stresses by comparing these genes against salt, cold, heat and oxidative stresses. The Gramene database was used in identification of sorghum genes associated with stress phenotypes based on known stress related ontology terms for each identified plant ontology. Ensembl BioMart was used to get sorghum orthologs having transitive association with known drought regulated functions from related species. The work flow provides a protocol for a step-by-step screening procedure to identify promising gene-sets for multiple stress tolerance across species: 1) The protocol identifies plant ontologies to query genes and detects if the genes belong to the sorghum gene association or to the orthologous group. Where there is no direct sorghum gene association, the protocol looks for orthologous group. Only those genes with these features were retained, and others were discarded. 2) The genes that were not supported by the relevant ontology terms in each ontology group were again rejected and only those with drought and associated ontology terms were screened for the next step. Once merged from all ontology groups, only unique genes were captured by removing the duplicates. 3) Among these, only those which were supported by all ontology groups were used for functional GO enrichment analysis and all others were discarded. 4) Functional GO enrichment analysis based on the P-value, FDR < 0.01 were used to screen the genes associated with stresses under investigation. Only those which satisfied this threshold value were selected as the candidates for the next step. 5) Comparative analysis across species and across traits was undertaken based on the above selected candidates. Sorghum specific and orthologous genes with multi-stress responses were combined with enrichment network and expression profiling for integrative analysis. Sorghum orthologs in other species were selected for which phylogenetic analysis was done. Key to legend: * Response to oxidative stress; ** Drought tolerance.