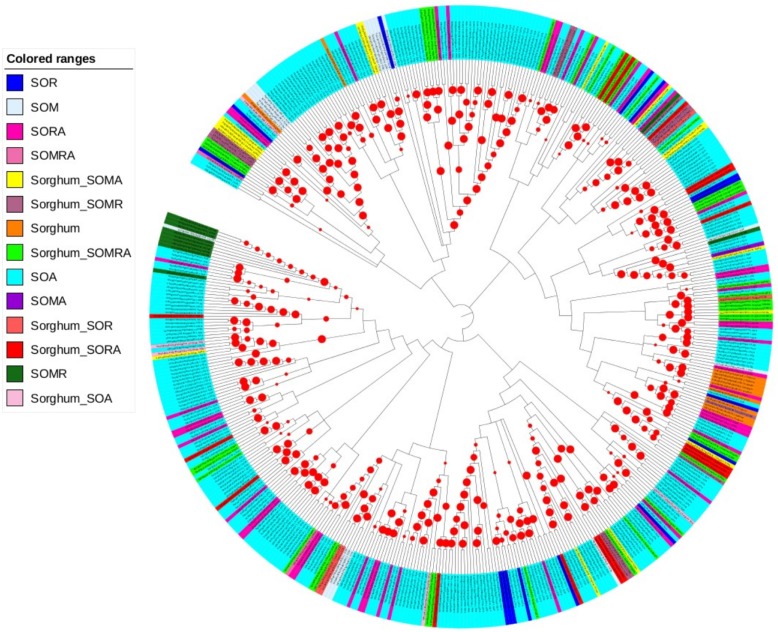
Fig 6. Circular representation of the polar formatted phylogenetic tree of the sorghum specific and orthologous genes.
Group of genes were color-coded by orthology group identified for drought response in the other species evolutionarily related to sorghum. The tree represents labels that were aligned with default leaf sorting. Branches represent evolutionarily related ortholog clades. Branch lengths for which 'ignored' setting was adjusted were represented each by the numbers in decimal and the bootstrap values in absolute numbers (S10 Table). The tree was reconstructed after removing the gaps using a bootstrap support of the 1,000 replicates to show the frequency of each internal node, clades in the tree. The red circular bootstrap symbol was used to indicate the bootstrap supported clades based on the values within the range of 100 (small dot)– 1000 (large dot) iterative replicates, where more than 75% of the clades showed the bootstrap above the commonly known threshold value (70%). The clades with the bootstrap values less than 5% were removed from the tree. The values for the robust bootstrap support were given in S10 Table. Key to legend for the colored ranges: SOA, Sorghum orthologs in Arabidopsis; SOM, sorghum orthologs in maize; SOMA, shared sorghum orthologs in maize and Arabidopsis; SOMR, shared sorghum orthologs in maize and rice; SOMRA, shared sorghum orthologs in maize, rice and Arabidopsis; SOR, sorghum orthologs in rice; SORA, shared sorghum orthologs in rice and Aabidopsis; Sorghum, sorghum specific genes; Sorghum_SOA, shared sorghum specific and sorghum orthologs in Arabidopsis; Sorghum_SOMA, shared sorghum specific and sorghum orthologs in maize and Arabidopsis; Sorghum_SOMR, shared sorghum specific and sorghum orthologs in maize and rice; Sorghum_SOMRA, shared sorghum specific and sorghum orthologs in maize, rice and Arabidopsis; Sorghum_SOR, shared sorghum specific and sorghum orthologs in rice; Sorghum_SORA, shared sorghum specific and sorghum orthologs in rice and Arabidopsis.