Figure 1. Using a culture-independent strategy for the discovery of calcium-dependent antibiotics from the global microbiome.
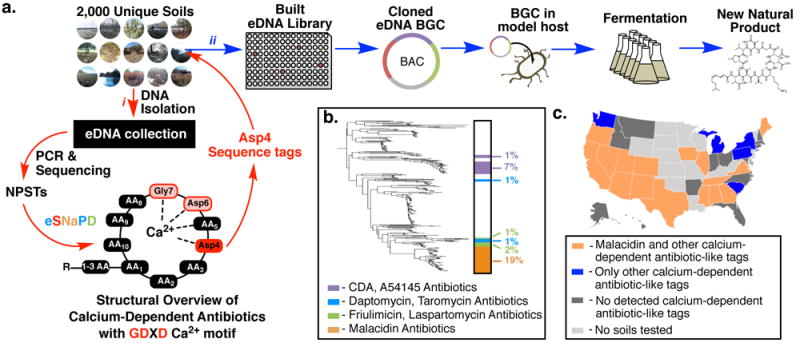
(a) i) Degenerate PCR primers targeting the conserved regions of adenylation domains (AD) found in nonribosomal peptide synthetase genes were used to generate amplicons from an arrayed collection of environmental DNA isolated from 2000 unique soils. The reads from these next-generation sequenced amplicons (natural product sequence tags, NPSTs) were analyzed by eSNaPD (environmental Surveyor of Natural Product Diversity). ii) A desert soil rich in AD-NPSTs from the previously unknown malacidin clade was used to build an arrayed cosmid library. Cosmids harboring all fragments of a targeted biosynthetic gene cluster were assembled and integrated into a heterologous host for production, extraction, and characterization. (b) AD-NPSTs identified by the eSNaPD analysis to be evolutionarily related to the conserved Asp4 ADs of known calcium-dependent antibiotics were used to phylogenetically map the unexplored clades of this larger family across all tested soil microbiomes. The subfamilies of calcium-dependent antibiotics and their relative abundance are illustrated on the phylogenetic tree by color and percent. Across all sampled soil metagenomes, the malacidin antibiotic-clade represents 19% of the NPSTs, and 59% of calcium-dependent antibiotic tags originate from unexplored branches. (c) Geospatial distribution of calcium-dependent antibiotics across sampled US soil metagenomes. US states containing at least one soil with AD-NPSTs from the malacidin clade are indicated in orange. States lacking malacidin tags but still containing calcium-dependent antibiotics NPSTs are indicated in blue. States with at least one sampled soil but no detected calcium-dependent antibiotics NPSTs are highlighted in dark grey.
