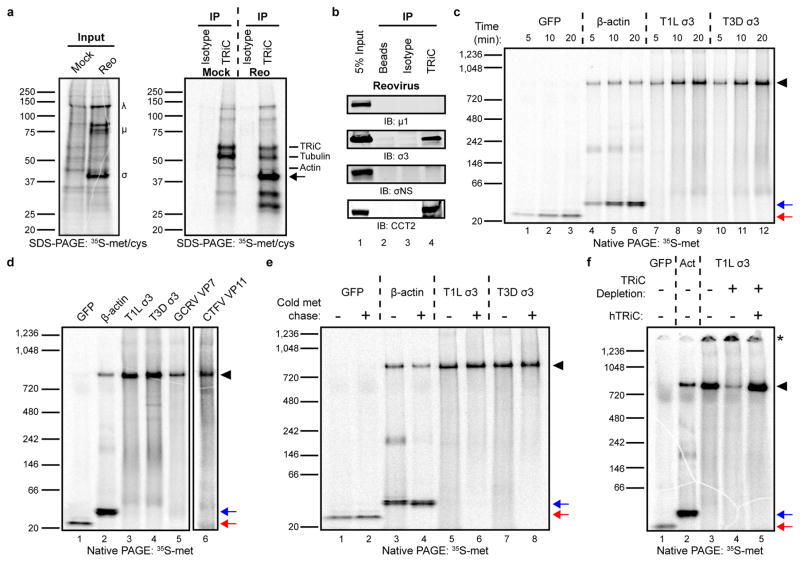
Figure 4. The TRiC chaperonin forms a complex with the reovirus σ3 outer-capsid protein.
a, SDS-PAGE of mock or T1L reovirus-infected HBMECs (MOI of 100 PFU/cell, 20 h post-infection) pulsed-labeled for 5 min with 35S-methionine/cysteine. Left gel: 2% input protein. Large (λ), medium (μ), and small (σ) viral proteins are labeled. Right gel: immunoprecipitation with isotype control or TRiC-specific antibody. Virus-specific band at ~40 kDa denoted by arrow. b, SDS-PAGE of TRiC immunoprecipitated from T3D reovirus-infected HBMECs (MOI of 100 PFU/cell, 48 h post-infection) immunoblotted for μ1, σ3, σNS, and CCT2. c, Native PAGE of 35S-methionine (met)-labeled GFP, β-actin, and T1L and T3D reovirus σ3 translated for the intervals shown in rabbit reticulocyte lysates (RRLs). d, Native PAGE of 35S-met-labeled GFP, β-actin, T1L and T3D σ3, GCRV VP7, and CTFV VP11 translated for 1 h in RRLs. CTFV VP11 is shown separately due low methionine content, requiring an independent exposure. e, Native PAGE of 35S-met-labeled GFP, β-actin, and T1L and T3D σ3 translated in RRLs with (+) or without (−) a 4 h cold methionine chase. f, Native PAGE of 35S-met-labeled GFP, β-actin (Act), and T1L σ3 translated in mock-depleted (−) or TRiC-depleted (+) RRLs with or without addition of purified human TRiC (hTRiC; +, 0.25 μM). The asterisk denotes protein unable to enter the native gel. In all native gels, the black arrowhead indicates TRiC-bound substrate, and the red and blue arrows indicate free GFP and β-actin monomers, respectively. In a-f, three independent experiments were conducted with similar results.