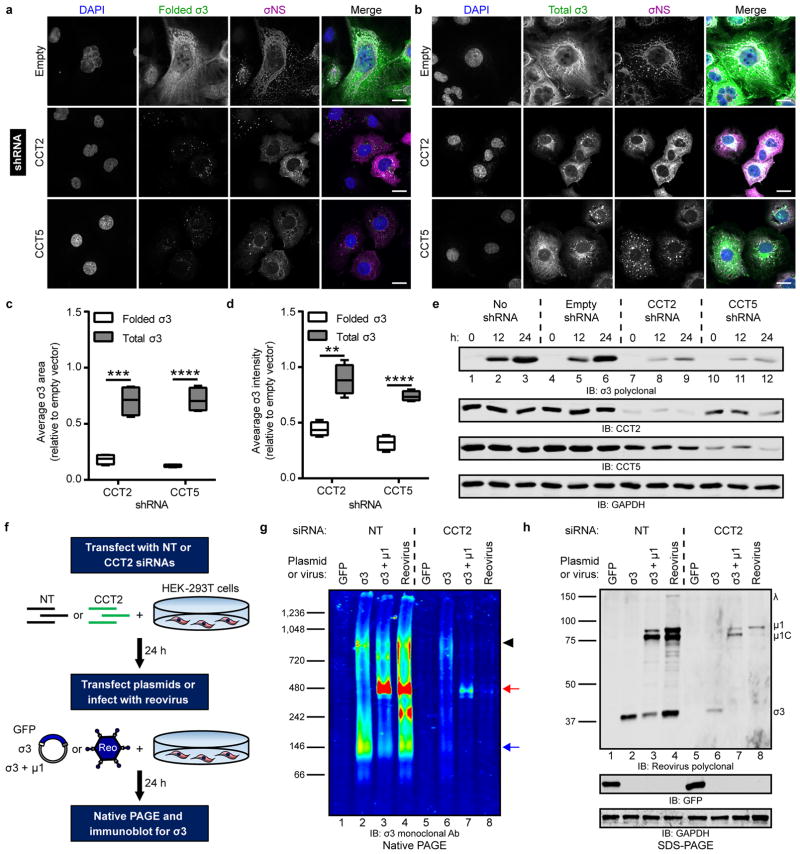
Figure 5. The intracellular biogenesis of native σ3 requires the TRiC chaperonin.
a,b, Immunofluorescence images of HBMECs stably transduced with TRiC subunit-specific shRNAs (or empty-vector), infected with T1L reovirus (MOI of 100 PFU/cell, 24 h post-infection), and stained with DAPI (blue) and antibodies specific for σNS (magenta) and folded (a, 10C1 monoclonal antibody) or total (b, VU219 polyclonal antiserum) σ3 (green). Scale bars, 20 μm. c,d, Quantification of folded or total σ3 staining area (c) or fluorescence intensity (d) per infected cell in CCT-shRNA transduced cells relative to empty vector control cells (> 200 cells quantified per experiment) from images acquired in (a) and (b). Data are presented as box-and-whisker plots of four independent experiments (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001, unpaired, two-tailed t-test). e, SDS-PAGE of shRNA-transduced HBMECs infected with T1L reovirus (MOI of 100 PFU/cell), harvested at the intervals shown, and immunoblotted for reovirus σ3, CCT2, CCT5, or GAPDH. f, Schematic of RNAi-knockdown and plasmid transfection or reovirus infection of HEK-293T cells. g, Native PAGE of HEK-293T cells transfected with a NT or TRiC subunit-specific siRNA and transfected with the indicated expression plasmids or infected with T3D reovirus (MOI of 100 PFU/cell, 24 h post-infection) and immunoblotted for σ3. Black arrowhead, TRiC-σ3 position; red arrow, μ13σ33 heterohexamer position; blue arrow, σ3 homo-oligomer position. h, SDS-PAGE of cell lysates from (g) immunoblotted for reovirus proteins, GFP, and GAPDH. Individual reovirus proteins are labeled. Immunoblots are representative of three independent experiments conducted with similar results.