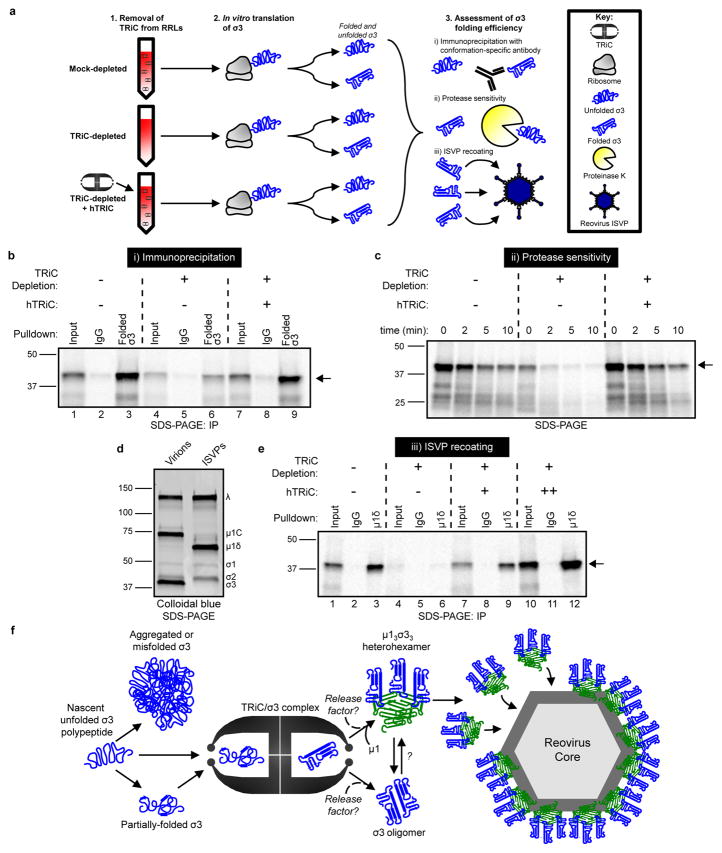
Figure 6. The TRiC chaperonin folds σ3 into a native, assembly-competent conformation.
a, Schematic of σ3 folding and assembly experiments. b, SDS-PAGE of 35S-met-labeled σ3 immunoprecipitated from RRLs using a conformation-specific antibody (10C1 monoclonal antibody) or isotype control. Where indicated, RRLs were TRiC-depleted and reconstituted with purified hTRiC (+, 0.25 μM) (10% input loaded into lanes 1,4,7). c, SDS-PAGE of 35S-met-labeled σ3 translated in RRLs mock-depleted, TRiC-depleted, or TRiC-depleted and reconstituted with purified hTRiC (+, 0.25 μM) and incubated with proteinase K (2.5 μg/mL final concentration) for the times shown. d, SDS-PAGE and colloidal blue stain of T1L reovirus virions and ISVPs. e, SDS-PAGE of ISVPs recoated with 35S-met-labeled σ3 translated in RRLs and immunoprecipitated using a μ1δ-specific antibody, which is specific to the μ1 species on ISVPs. Where indicated, RRLs were TRiC-depleted and reconstituted with purified hTRiC (+, 0.125 μM; ++, 0.50 μM) (20% input loaded into lanes 1,4,7,10). Arrows in SDS-PAGE gels indicate full-length σ3. In b–e, three independent experiments were conducted with similar results. f, Model of the TRiC-σ3 folding and assembly pathway.