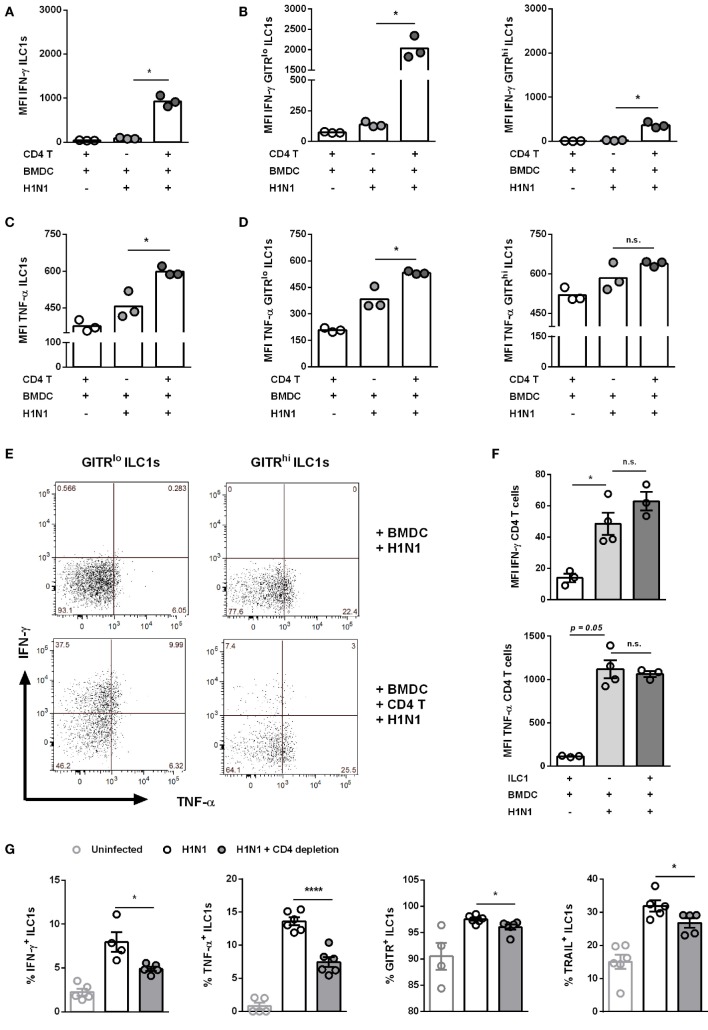
Figure 8.
CD4 T cells contribute to enhanced ILC1 functionality during H1N1 infection in vitro and in vivo. FLT-3 ligand-differentiated BMDCs were infected with the OVA peptide (aa323–393)-expressing H1N1 PR8 influenza strain at a MOI of 1. In vitro-generated ILC1s were cocultured overnight with infected or uninfected BMDCs and CD4 T cells derived from OT II mice at a 1:1:1 ratio. Staining of surface makers and cytokines for flow cytometry analysis after 3 h of incubation in media with brefeldin and monensin. MFI of (A,B) IFN-γ and (C,D) TNF-α expression by ILC1s depending on the level of GITR expression. (E) Representative two-dimensional FACS plots of IFN-γ vs. TNF-α from GITRlo and GITRhi ILC1s in the absence or presence of CD4 T cells. (F) MFI of IFN-γ and TNF-α-expressing CD4 T cells. CD4 T cells were depleted before and 3 days after H1N1 PR8 infection (2 × 103 ffu/animal) of wild-type mice by i.p. administration of CD4 T cell-depleting antibodies (200 µg/animal). Lung-derived lymphocytes of infected wild-type mice were stained for flow cytometry analysis 6 dpi with regard to markers related to ILC1 activation and functionality. (G) Frequencies of IFN-γ+, TNF-α+, TRAIL+, and GITR+ lung-derived ILC1s. Bars with scatter plots represent the mean ± SEM and the in vitro (n = 3–4 technical replicates) and in vivo (n = 5–6) data are representative from one out of two independent experiments. Asterisks denote significant values as calculated by nonparametric Kruskal–Wallis test (Dunn’s posttest) (in vitro data) or One-way ANOVA (in vivo data); ****p ≤ 0.0001; ***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05; n.s., not significant.