ABSTRACT
There are seven conserved CTCF binding domains in the herpes simplex virus 1 (HSV-1) genome. These binding sites individually flank the latency-associated transcript (LAT) and the immediate early (IE) gene regions, suggesting that CTCF insulators differentially control transcriptional domains in HSV-1 latency. In this work, we show that two CTCF binding motifs in HSV-1 display enhancer blocking in a cell-type-specific manner. We found that CTCF binding to the latent HSV-1 genome was LAT dependent and that the quantity of bound CTCF was site specific. Following reactivation, CTCF eviction was dynamic, suggesting that each CTCF site was independently regulated. We explored whether CTCF sites recruit the polycomb-repressive complex 2 (PRC2) to establish repressive domains through a CTCF-Suz12 interaction and found that Suz12 colocalized to the CTCF insulators flanking the ICP0 and ICP4 regions and, conversely, was removed at early times postreactivation. Collectively, these data support the idea that CTCF sites in HSV-1 are independently regulated and may contribute to lytic-latent HSV-1 control in a site-specific manner.
IMPORTANCE The role of chromatin insulators in DNA viruses is an area of interest. It has been shown in several beta- and gammaherpesviruses that insulators likely control the lytic transcriptional profile through protein recruitment and through the formation of three-dimensional (3D) chromatin loops. The ability of insulators to regulate alphaherpesviruses has been understudied to date. The alphaherpesvirus HSV-1 has seven conserved insulator binding motifs that flank regions of the genome known to contribute to the establishment of latency. Our work presented here contributes to the understanding of how insulators control transcription of HSV-1.
KEYWORDS: insulator, CTCF, HSV-1 reactivation, epigenetics, chromatin, mouse ocular, in vivo reactivation, PRC2, Suz12, herpes simplex virus
INTRODUCTION
Herpes simplex virus 1 (HSV-1) establishes a lifelong latent infection in host sensory neurons and is a significant human pathogen due to its ability to exit latency and reactivate to cause clinical disease (1, 2). The primary infection occurs in the mucosal epithelium, where the virus enters into the cell and is organized into dynamic nucleosomes that regulate lytic gene transcription (3). Following the establishment of latency in host sensory neurons, lytic gene transcription is essentially silenced, and the latency-associated transcript (LAT) is the only transcript abundantly expressed (4). The primary LAT is an 8.5-kb noncoding RNA (ncRNA), and the 5′ exon region of the LAT contains the LAT promoter and LAT enhancer, which together are deemed the reactivation-critical region of LAT (5–7). Further, LAT expression likely plays a pivotal role in the establishment and maintenance of latency by regulating the lytic transcriptional program (8–10).
The latent genome exists as a circular episome associated with histones that bear histone marks corresponding to the transcriptional status of genes. For example, both the LAT promoter and enhancer are enriched in the transcriptionally active histone marks acetyl H3K9K14 (histone H3 acetylated at K9 and K14) and H3K4me2 (histone H3 dimethylated at K4) during latency (11–13). In contrast, the juxtaposed immediate early (IE) lytic genes of ICP0, ICP4, and ICP27 are predominantly enriched in the transcriptionally repressive facultative heterochromatic histone mark H3K27me3 (histone H3 trimethylated at K27) during latency (9, 14–18). LAT transcription was shown to downregulate lytic gene transcription during latency through the deposition of repressive histone marks on the IE lytic promoters. In the absence of LAT, repressive histone marks on these lytic promoters were less abundant (9, 10, 14), and these findings were consistent with the observation that leaky lytic transcription occurs in LAT-null mutants (19). Additional reports showed that facultative heterochromatic marks differentially increased on the ICP4 promoter in the 17ΔPst virus, a mutant with a 202-bp deletion of the core promoter, and indicated that LAT transcription played a role in directing the dynamic chromatinization of the HSV-1 genome during latency to allow for reactivation (8). While it is clear that LAT expression is critical for the establishment and maintenance of latency, the mechanism by which this occurs has not yet been established. However, the organization of the LAT and IE transcriptional domains in latent HSV-1 indicates that chromatin insulator elements may be key functional components in the control of gene expression.
Chromatin insulators are long-range cis-acting DNA sequence elements that have the ability to regulate transcriptional domains and gene expression by preventing inappropriate signaling from one transcriptional domain to another (20). Predominantly characterized in eukaryotes, chromatin insulators consist of the insulator protein bound to the DNA element. The most common insulator protein is CTCF, an 11-zinc-finger nuclear protein that regulates transcriptional domains through a number of functions, including promoter activation, repression, silencing, and barrier activity (21). Previous sequence analysis of the HSV-1 genome identified seven conserved, reiterated CTCF binding motifs during latency (22). Provocatively, these binding domains flank the reactivation-critical region of the LAT and each individual IE gene region (22), indicating that chromatin insulators may regulate individual gene regions in HSV-1. We subsequently showed that three of these CTCF binding sites were occupied by CTCF during latency, including the CTRL2 site downstream from the LAT enhancer, CTa′m upstream from ICP0, and CTRS3 upstream from ICP4 (Fig. 1). All three sites were also characterized as functional insulators with enhancer-blocking ability in rabbit skin cells (22, 23). Finally, we showed that CTCF was evicted from DNA binding sites at very early times postreactivation in vivo in mice latently infected with the wild-type (wt) virus 17Syn+ but not in mutants that lacked the ability to reactivate, indicating that CTCF eviction was also an important component of reactivation (23).
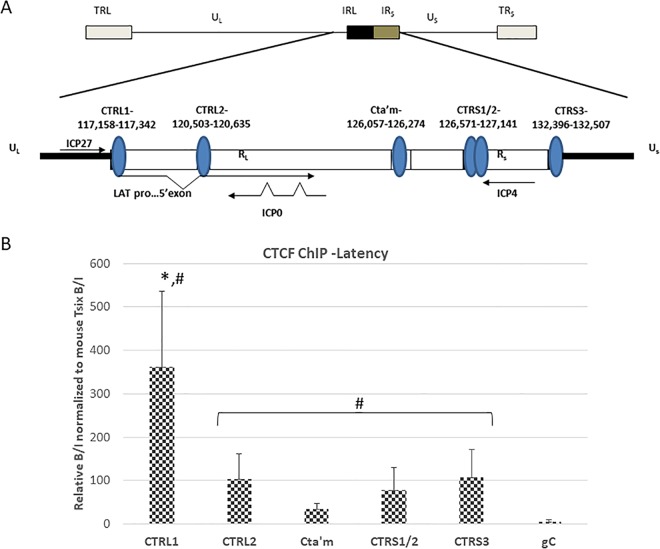
FIG 1.
CTCF occupation of the CTCF binding motif in HSV-1 during latency in mouse TG. (A) Genomic positions of CTCF binding motifs in the HSV-1 genome. It should be noted that CTRS1/2 is actually two CTCF binding clusters separated by less than 100 nucleotides. There is an additional CTCF motif (CTUS1) identified by Amelio et al. (22) not shown in this figure. (B) ChIP data are presented as a ratio of the relative copy numbers of the PCR target in two fractions (bound and input [B/I]), where the bound fractions are representative of aliquots incubated overnight with anti-CTCF, and the input fractions are representative of the total DNA present. Relative copy numbers in the B or I fraction were determined from the equation for the standard curve specific to the primer/probe set used. ChIP assays were validated by determining the B/I ratios of the cellular controls Tsix imprinting/choice center CTCF site A (positive control) and MT498 (negative control). Each ChIP assay was done using pooled samples from three mice (6 TG pooled) (n = 8). All relative B/I ratios from each gene region were normalized to the B/I ratio for the cellular control Tsix site A for that ChIP assay. Statistical data were determined by one-way analysis of variance by comparing the B/I ratios of each gene region to the B/I ratio for the CTRL1 site (*, P < 0.01) as well as to the negative control, gC (#, P < 0.05). UL, unique long region; US, unique short region; RL, repeat long region; RS, repeat short region.
The ability of CTCF insulators to recruit and interact with both coactivator and corepressor proteins is important for the regulation of gene expression in mammalian cells. For example, it was shown that a subpopulation of CTCF interacted with RNA polymerase (Pol) II to activate transcription of a reporter gene (24). Other reports showed that CTCF recruited the corepressor proteins Sin3 and YB1 to repress the transcription of the c-myc promoter (25), and CTCF directly interacted with Suz12, one of the five proteins that comprise polycomb repressive complex 2 (PRC2) (26). PRC2 trimethylates histone 3 at lysine 27 (H3K27me3) to silence transcription and in mammalian cells, a CTCF-Suz12 interaction promoted allele-specific deposition of H3K27me3 on maternal Igf2 promoters to repress expression and control Igf2 imprinting (26). The PRC2 is an important epigenetic regulating complex comprised of five proteins (EZH2, SUZ12, EED, and RdAp46/48) and is responsible for the trimethylation of H3K27 (H3K27me3) to silence gene transcription (27). The fact that HSV-1 IE lytic regions are populated with the heterochromatic histone mark H3K27me3 during latency indicates that a corepressor protein may be recruiting PRC2 for genome maintenance. In support of this, Cliffe et al. recently showed that the PRC2 protein Suz12 was recruited to the LAT promoter, ICP8, and UL48 following the resolution of acute infection in mice (postinfection [p.i.] day 14) in a LAT-independent manner but not to the ICP0 and ICP4 promoters (14), suggesting that other elements may be directing PRC2 recruitment for the deposition of H3K27me3 to these sites to repress them during latency. We hypothesize that CTCF bound to the sites flanking the IE genes interacts with Suz12 in the PRC2 to establish latency. In this work we show that CTCF enrichment, protein recruitment, and insulator function at the individual HSV-1 sites were independent and site specific. We also show that the insulators CTRL1 and CTRS3 have different enhancer-blocking abilities depending on the cell type (neuronal or epithelial). This finding indicates that these sites have different insulator activities during the lytic and latent stages of infection. We found that Suz12 colocalizes predominantly to CTCF sites flanking the IE genes of ICP0 and ICP4 and not to these IE promoters, indicating that CTCF and Suz12 act as corepressors that together establish repressive domains during latency by a Suz12-CTCF interaction. This finding is further supported by our data showing that Suz12 enrichment significantly decreases at the IE sites following reactivation.
RESULTS
The CTRL1 site is significantly more enriched in CTCF during latency.
In 2006, Amelio et al. used sequence analysis of the wt 17Syn+ latent genome and identified seven conserved reiterated consensus binding motifs for the insulator protein CTCF (22). We subsequently showed that three of these identified sites, the CTRL2, CTa′m, and CTRS3 sites (located downstream of the LAT enhancer and upstream of the ICP0 and ICP4 promoters, respectively) (Fig. 1A) were enriched in CTCF during latency in mouse trigeminal ganglia (TG) following an ocular route of infection (23). We hypothesized that individual CTCF sites may be differentially regulated, recruit different coregulators to control lytic gene transcription, and have different insulator functions. To further explore this hypothesis, we quantified CTCF binding on two additional CTCF binding motifs in HSV-1, specifically the CTRL1 site upstream of the LAT promoter (nucleotides [nt] 117159 to 117342), which segregates ICP27 from the LAT promoter/enhancer elements, and the CTRS1/CTRS2 (CTRS1/2) combination site (nt 126571 to 127141), which is downstream of ICP4 (Fig. 1A) (22). Prior to PCR, all chromatin immunoprecipitations (ChIPs) were validated using cellular controls as described in Materials and Methods. All ChIP data are presented as normalized to the cellular control mouse gene Tsix. Mice were ocularly infected with the wild-type virus 17Syn+, and a latent infection was established in the mice (28 days p.i.). We harvested the TG and performed ChIP assays combined with quantitative PCR (qPCR) to quantify enrichments of CTCF bound to the CTRL1 and CTRS1/2 sites. Here, we found a significant enrichment of CTCF bound to the CTRL1 site (>3-fold; P < 0.01), relative to all other CTCF binding domains in the latent genome (all of these sites are significantly enriched in CTCF relative to the level in the negative control, gC) (Fig. 1B). Collectively, our data suggest that CTCF occupation of each individual site is differential.
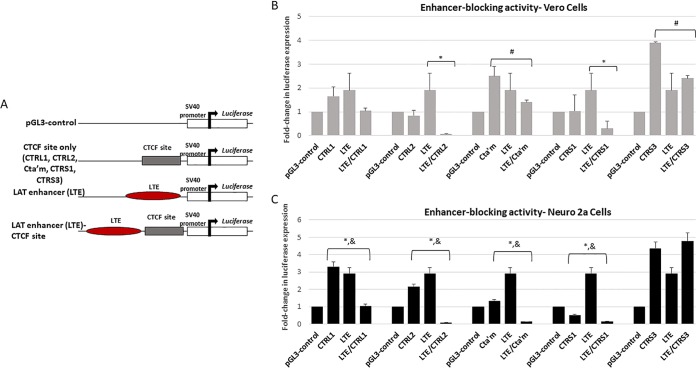
The CTRL1 site and the CTRS3 site have cell-type-specific enhancer-blocking activity.
The CTRL2, CTa′m, and CTRS3 sites were previously shown to be insulators that have the ability to block the LAT enhancer in rabbit skin cells (22, 23). Here, we sought to determine if enhancer blocking was consistent among all CTCF sites characterized in HSV-1 in reporter assays with epithelial cells. To test the ability of the CTRL1, CTRS3, and the CTRS1/2 sites to block enhancer activity, we generated reporter constructs using a commercially available pGL3 control plasmid and tested each construct for enhancer-blocking function using standard luciferase assays in transient transfections in Vero cells (23). These reporter constructs were generated to include the core CTCF motif together with 300 to 1,200 bp of site-specific nucleotides upstream and downstream of the core CTCF motifs since functional properties of eukaryotic insulator elements are often located outside the CTCF binding sites (20). In addition, we used CTRL2 and CTa′m constructs, together with the corresponding long-term expression element (LTE)/CTRL2 and LTE/CTa′m plasmids for direct comparison in these reporter assays (Fig. 2A shows a schematic representation of the constructs). The results for our reporter constructs tested in Vero cells are presented in Fig. 2B. Interestingly, the CTRL1 reporter construct did not act as a functional enhancer blocker in Vero cells as we observed no significant decrease in luciferase expression levels between the LTE construct and the CTRL1/LTE reporter (Fig. 2B). By comparison, only the CTRL2 and CTRS1 reporters displayed enhancer-blocking capability toward the LTE in Vero cells (Fig. 2B). Baseline luciferase expression levels for both the CTa′m and the CTRS3 reporters were on average 2.5- to 4-fold higher than the level of the control (pGL3) reporter in Vero cells, and so additional comparisons of the CTCF and LTE/CTCF constructs were done to determine if a decrease in luciferase expression could be observed between these reporter constructs at the corresponding LTE/CTCF reporters. By directly comparing the CTa′m and LTE/CTa′m or the CTRS3 and LTE/CTRS3 reporters, we found a decrease in luciferase expression (∼2-fold) (Fig. 2B, #), indicating that these sites may have weak enhancer-blocking activity in Vero cells, consistent with previous reports showing that each site was a functional enhancer blocker in rabbit skin cells (12). Nevertheless, these data confirm that the CTRL1 binding site of HSV-1 is not a functional enhancer blocker in epithelial cells, indicating that CTRL1 may have an alternative insulator function not yet identified. In eukaryotic gene expression, CTCF insulators have different cell-type-specific insulator functions and can recruit different coregulatory proteins in response to cell type. To determine whether the CTCF binding motifs identified in the latent HSV-1 genome also have cell-type-specific insulator function, we tested these reporter constructs in a neuronal cell line. When we tested the CTRL1 plasmids in reporter assays using Neuro 2a cells, we found that the CTRL1 site was a potent enhancer blocker of the LAT enhancer, with luciferase expression decreased by 5-fold in the CTRL1/LTE construct compared to the levels of the LTE and the CTRL1 constructs (Fig. 2C). Similar results were found for the CTRL2, CTa′m, and CTRS1 sites of HSV-1, where significant decreased luciferase expression was observed in the LTE/CTCF reporters compared to levels of both the LTE and the respective CTCF reporters (Fig. 2C). However, when we tested the ability of the CTRS3 site to block the LAT enhancer in Neuro 2a cells, we found that this site was not a functional enhancer blocker in neuronal cells, in contrast to the enhancer-blocking activity observed in other epithelial cell lines (Fig. 2C) (23). These striking cell-type-specific differences in insulator phenotypes indicate that the CTRL1 and CTRS3 sites have dynamic insulator functions during lytic (epithelial cells) and latent (neuronal cells) infections.
FIG 2.
Luciferase reporter constructs were generated to test the ability of the CTCF binding motif to block the LAT enhancer. (A) General schematic for construct design. Each CTCF/LTE construct contained the specific CTCF binding site positioned between the LAT enhancer element and the simian virus 40 (SV40) promoter in the pGL3 control vector. (B) Reporter assays in Vero cells. All transfections were done in triplicate wells and were repeated five times (n = 5). Data are presented as both a comparison of the LTE to the LTE/CTCF construct and of the CTCF compared to the LTE/CTCF construct (*, P < 0.001), comparing the reduction in luciferase expression levels between the LTE and LTE/CTCF constructs, determined by unpaired one-tailed Student's t tests in pairwise comparisons; #, 2-fold reduction in luciferase expression levels between the CTCF and LTE/CTCF constructs. All luciferase values were normalized to the value for the pGL3 control, which was set to 1. (C) Reporter assays done in Neuro 2a cells. All transfections were done in triplicate wells and were repeated five times (n = 5). Data are presented as both a comparison of the LTE to the LTE/CTCF construct and of the CTCF compared to the LTE/CTCF construct (*, P < 0.001), comparing the reduction in luciferase expression levels between the LTE and LTE/CTCF constructs, determined by unpaired one-tailed Student's t tests in pairwise comparisons; &, P < 0.01, comparing the reduction in luciferase expression levels between the CTCF and LTE/CTCF constructs.
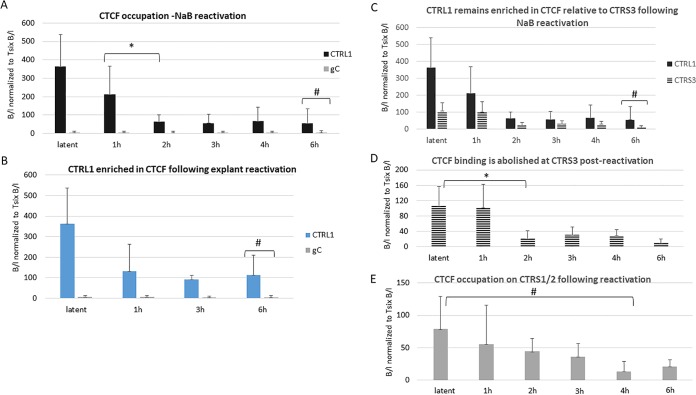
CTCF is not completely evicted from the CTRL1 site at early times postreactivation.
We hypothesized that CTCF eviction might be a fundamental step in the process of reactivation and previously showed that CTCF was completely evicted from the CTa′m and CTRS3 sites upstream of the lytic promoters from the latent HSV-1 genome at early times following reactivation (23). To determine whether the CTRL1 and CTRS1/2 sites also evict CTCF at early times postreactivation, we utilized two methods of reactivation, sodium butyrate (NaB) treatment and explant-induced reactivation. Following NaB treatment, the mouse TG were harvested at 1 h, 2 h, 3 h, 4 h, and 6 h. ChIP assays combined with real-time PCR analyses of the TG from NaB-treated mice showed that by 2 h post-NaB treatment, the CTRL1 lost CTCF enrichment relative to the latent time point but remained significantly enriched in CTCF, relative not only to the negative control (gC) (Fig. 3A) but also to other CTCF binding sites, such as the CTRS3 site, where CTCF binding is essentially abolished by 6 h post-NaB treatment (Fig. 3C and D); this suggests that the CTRL1 region may retain insulator capabilities during a reactivation event. To ensure that the changes being observed with respect to CTCF eviction were a consequence of reactivation and not a direct result of NaB treatment, we confirmed our results using an alternative method of HSV-1 reactivation in the mouse. Here, we employed the explant-induced HSV-1 reactivation model, an ex vivo model that has been consistently and reliably used to study HSV-1 reactivation in latently infected mice (28, 29). We harvested mouse TG latently infected with wild-type 17Syn+, placed them in supplemented medium, and incubated them at 37°C with 5% CO2 for 1, 3, or 6 h, as previously reported (28). ChIP assays were performed as described for NaB experiments. The results from our explant-induced reactivation ChIP experiments were consistent with the NaB experiments and confirmed that CTCF enrichment decreased postexplant (Fig. 3B). We also examined the CTRS1/2 sites using both NaB (Fig. 3E) and explant-induced reactivation (data not shown). Here, we observed CTCF eviction with the CTRS1/2 site, consistent with the CTCF eviction from the CTRL2, CTa′m, and CTRS3 sites following reactivation (23). It is important to note that we also used PCR with HSV-1 DNA Pol primers to quantify HSV-1 genomes in ganglia during latency and following reactivation at the time points listed in Fig. 3, and we found no significant difference in the number of genomes present, proving that the CTCF eviction observed was not due to changes in genome loads following reactivation (data not shown). These data further support that CTCF eviction during reactivation is dynamic and site specific, with the CTRL1 site retaining bound CTCF following reactivation.
FIG 3.
CTRL1 remains enriched in CTCF following reactivation. (A) Mice latently infected with HSV-1 17Syn+ were subjected to intraperitoneal treatment with sodium butyrate (NaB), and the TG were harvested at 1 h, 2 h, 3 h, 4 h, and 6 h. ChIP assays using anti-CTCF were combined with real-time PCR analyses to compare TG from NaB-treated mice and latently infected mouse TG at the CTRL1 site. Time following NaB treatment is represented on the x axis. The 0-h time point samples are from latently infected mice not subjected to NaB treatment. TG of three mice (6 TG) were pooled for each ChIP assay (n = 8 for each time point done). There was a significant decrease in CTCF occupation of this site by 2 h post-NaB treatment (>3-fold; *, P < 0.01), but the site remained significantly enriched in CTCF relative to the level of the negative control, gC (#, P < 0.05) (B) Explant-induced reactivation was done to confirm NaB results. TG of three mice (6 TG) were pooled for each ChIP assay (n = 8 for each time point done). Data are consistent with results for NaB-treated animals where CTCF is not completely evicted from the CTRL1 site (#, P < 0.05, one-way ANOVA). (C) CTRL1 is significantly more enriched than CTRS3 at 6 h postreactivation following NaB (#, P < 0.05). (D) CTCF binding at the CTRS3 site is significantly lower by 2 h (*, P < 0.01) and is essentially abolished by 6 h postreactivation. (E) CTCF is significantly lower at the CTRS1/2 sites by 4 h following NaB reactivation (#, P < 0.05). Statistical data were determined by one-way ANOVA by comparing the B/I ratios at each time point shown. It should be noted that latent samples are the same samples used in the experiment shown in Fig. 1B.
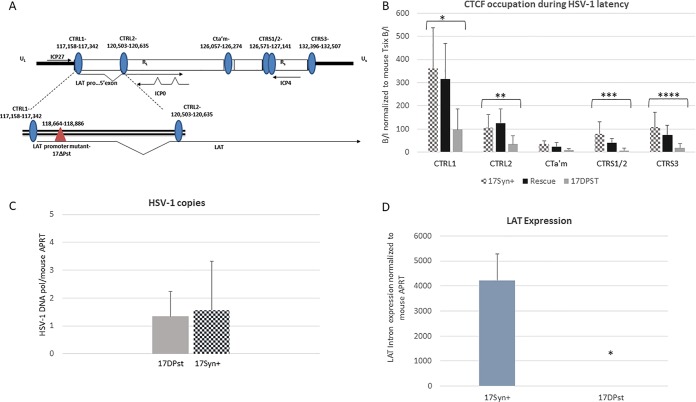
CTCF occupation on the latent genome is differentially driven by LAT.
LAT expression may play a pivotal role in the establishment and maintenance of latency through regulation of lytic gene expression. To determine whether LAT contributes to the regulation of gene expression through a CTCF-mediated mechanism, we examined CTCF occupation on the latent HSV-1 genome in a LAT promoter mutant virus, 17ΔPst (the deletion lies outside the CTRL1 binding motif) (Fig. 4A). Deletion of the 202-bp core promoter of LAT results in >10,000-fold reductions in LAT expression during latency in vivo (Fig. 4D). ChIP assays using anti-CTCF were performed using mouse TG harvested at 28 days postinfection. Again, ChIP assays were validated using cellular controls as described in Materials and Methods, and all viral targets were normalized to the host mouse Tsix ratios so that comparisons could be made between viruses. Using qPCR, we found that CTCF binding was significantly lower on the CTRL1, CTRL2, CTa′m, and CTRS3 sites of the genome in the LAT-null mutant 17ΔPst than of the wild-type virus and the rescued 17ΔPst virus (Fig. 4B). Comparison of the wild-type, rescued, and mutant viruses show that there is a significant decrease in CTCF enrichment at all CTCF binding sites in the LAT-null mutant at all sites except the CTa′m site, upstream of the ICP0 promoter region of HSV-1, where CTCF enrichment is the lowest in wt virus (Fig. 4B). Further, the decrease in CTCF binding in the mutant is not the result of either compensatory changes in the conserved DNA binding sequence or the deletion itself. We observed no significant change in the CTCF enrichment of any sites in the rescue of 17ΔPst. Finally, the decreased CTCF enrichment observed in the mutant was not due to decreased genome loads between the mutant and wild-type viruses as equivalent infections were observed in both sets of ganglia (Fig. 4C). Taken together, these data indicate that abundant CTCF occupancy on the HSV-1 genome during latency is linked with the ability to express LAT.
FIG 4.
CTCF occupation is LAT dependent. The bar graphs represent ChIP assays using anti-CTCF as previously described. For each virus TG of three mice (6 TG) were pooled in one sample (n = 8). Relative enrichments for each gene region are shown for all three viruses. Statistical analyses were done for each site comparing the wt, mutant, and rescue samples. (A) Diagram of the deletion relative to the CTRL1 site in HSV-1. (B) In the LAT-null mutant virus 17ΔPst, CTCF occupation of each site is significantly lower at all sites, excluding CTa′m (for all comparisons, asterisks indicate a P value of <0.05), relative to the levels of wild-type and rescue viruses. (C) The number of HSV-1 genomes per sample was determined to show equivalent infections among the wt and mutant viruses. Relative copy numbers of HSV-1 DNA polymerase and mouse APRT were determined by PCR using 8 mice (16 TG) that were latently infected with 17Syn+ or 17ΔPst. TG were harvested at 28 days postinfection, and DNA was extracted. Data are presented as a ratio of HSV-1 DNA Pol/APRT. One-way ANOVA did not indicate a significant difference in HSV-1 copy numbers between the viruses. (D) Decreased LAT expression for the 17ΔPst virus was confirmed by qRT-PCR using primers/probe sequences specific for the LAT intron and normalized to mouse APRT (n = 8). Mice ganglia latent with either wt or 17ΔPst virus were harvested, and RNA was extracted as described in Materials and Methods.
Suz12 colocalized to CTCF insulators flanking the IE regions of HSV-1 during latency.
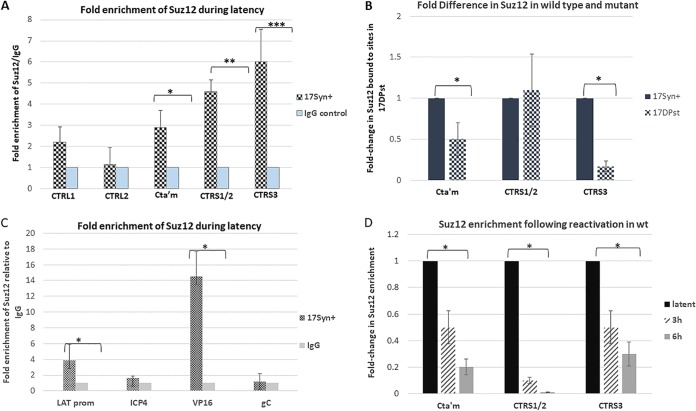
In mammalian genomes, it was demonstrated that CTCF interacted with the protein Suz12 in the PRC2 complex, leading to allele-specific H3K27 methylation to suppress maternal Igf2 promoters for gene silencing. Further, Cliffe et al. subsequently showed that Suz12 is recruited to specific lytic promoters in a LAT-independent manner following the resolution of the acute infection at p.i. day 14 but not to ICP0 and ICP4 promoters (14). In our experiments, we sought to determine if CTCF interacted with Suz12 to potentially recruit the PRC2 complex to insulator sites for transcriptional control. We performed ChIP assays using anti-Suz12 combined with real-time PCR of the individual CTCF binding motifs CTRL1, CTRL2, CTa′m, CTRS1/2, and CTRS3 to determine whether Suz12 was recruited to these sites that are enriched in CTCF during latency in wild-type virus. We compared the ratio of bound to input (B/I) viral DNA precipitated from the ChIP antibody for Suz12, with the B/I value of viral DNA precipitated with the nonspecific IgG antibody as a negative control, and the data are presented as the fold-enrichment of Suz12 compared to the IgG level (Fig. 5A). We found evidence that Suz12 was recruited to the CTa′m, CTRS1/2, and CTRS3 sires that flank the ICP0 and ICP4 gene regions of HSV-1 but not to the CTRL1 and CTRL2 sites around the LAT region (Fig. 5A). Our data indicate that CTCF bound to these insulator sites may facilitate the recruitment of the PRC2 through a direct Suz12-CTCF interaction to repress lytic IE transcription during latency. In support of this, ChIP assays done to quantify Suz12 on the CTa′m, CTRS1/2, and the CTRS3 sites in the mutant 17ΔPst show a significant decrease in Suz12 on the CTa′m and CTRS3 sites relative to wt levels (Fig. 5B). Since there is less CTCF bound to these sites in the mutant virus, the decreased enrichment of CTCF at these sites in 17ΔPst further support the hypothesis that Suz12 is recruited by CTCF. Additional qPCR was done on wt virus following the Suz12 ChIP assays to confirm that Suz12 localized to the LAT promoter and VP16 sites, as previously reported by Cliffe et al. (14). We found a significant enrichment of Suz12 on both of these sites (Fig. 5C), indicating that the PRC2 complex might be recruited by other corepressor elements in these regions. In contrast, we found no significant enrichment of Suz12 on the ICP4 promoter region. This, combined with the fact that the ICP4 promoter is over 1,000 bp away from the nearest CTCF site (CTRS3), led us to conclude that Suz12 and CTCF interact at the CTCF binding site and not the ICP4 promoter. We also assayed by ChIP the enrichment of Suz12 on the CTCF sites of HSV-1 at early times post-NaB reactivation. Here, we found that Suz12 enrichment significantly decreased by 6 h postreactivation on the sites flanking the IE regions of ICP0 and ICP4 (Fig. 5D). Further, this time frame for the reduction of Suz12 is consistent with both CTCF eviction and changes to the histone marks of these sites, further supporting the idea that CTCF recruits the PRC2 through a Suz12-CTCF interaction to maintain IE repression during latency. Conversely, no changes could be observed in Suz12 enrichment at the CTRL1 or CTRL2 sites following reactivation (data not shown for simplicity).
FIG 5.
Suz12 is recruited to CTCF binding sites flanking ICP0 and ICP4. The bar graph represents ChIP assays using anti-Suz12 for the wt virus and for comparison of wt and mutant viruses. qPCR was done on individual CTCF binding motifs of CTRL1, CTRL2, CTa′m, CTRS1/2, and CTRS3 following ChIP assays using anti-Suz12 (Millipore). (A) Nonspecific IgG antibody was used as a control for binding in the bound samples of each ChIP assay (n = 8). Data are presented as the fold enrichment of Suz12 compared to the IgG level, where relative values for IgG were set to 1. Error bars represent the standard deviations between each fold change for each individual ChIP assay done. There was a significant enrichment of Suz12 at the CTa′m, CTRS1/2, and CTRS3 sites relative to the level of the antibody control, IgG (*, P < 0.05; **, P < 0.01; ***, P < 0.01). (B) ChIP assays were done comparing Suz12 enrichment at the CTa′m, CTRS1/2, and CTRS3 sites wt and mutant viruses (n = 6). Data are presented as fold change in Suz12 enrichment at each site for 17ΔPst. The wt virus (17Syn+) enrichment for each site was normalized to 1. There were significant decreases in the enrichments of the CTa′m and CTRS3 sites for 17ΔPst relative to wt virus (*, P < 0.05). (C) ChIP assays using the Suz12 antibody followed by qPCR of viral targets, including the LAT promoter, ICP4, and VP16, were done. gC was used as a negative viral control. We found significant enrichment of Suz12 on the LAT promoter and VP16 sites (*, P < 0.05) but not on ICP4. Nonspecific IgG antibody was used as a control for binding in the bound samples of each ChIP assay (n = 8). Data are presented as the fold enrichment of Suz12 compared to the IgG level, where relative values for IgG were set to 1. Error bars represent the standard deviations between each fold change for each individual ChIP assay done. Statistical data were determined by one-way analysis of variance by comparing the B/I ratios of the viral targets to the ratio of the IgG control. (D) The bar graph represents ChIP assays using anti-Suz12 for the wt virus following NaB reactivation, as described in Materials and Methods. qPCR was done on individual CTCF binding motifs following ChIP assays using anti-Suz12 (Millipore). Nonspecific IgG antibody was used as a control for binding in the bound samples of each ChIP assay (n = 8). Data are presented as the fold enrichment of Suz12 at each site during latency, where relative values for Suz12 bound/input and IgG bound/input were set to 1. Error bars represent the standard deviations between each fold change value for each individual ChIP assay done. There was a significant decrease in enrichment of Suz12 at the CTa′m, CTRS1/2, and CTRS3 sites by 6 h postreactivation (*, P < 0.05).
DISCUSSION
The hypothesis that CTCF insulators regulate the establishment and maintenance of transcriptional domains in HSV-1 latency is very provocative. Because HSV-1 lytically replicates in epithelial cells but establishes latency in neurons, we hypothesized that the CTCF insulators may function differently depending on the cell type. Therefore, we generated reporter constructs and tested these constructs in Vero cells (epithelial) and Neuro 2a cells as a model for neurons. We showed that the CTRL1 site does not function as an enhancer blocker in epithelial cells but displays a potent enhancer-blocking function in neuronal cells. This finding is consistent with another report showing that elements in the CTRL1 motif have cell-type-specific functions in neurons and fibroblasts (30). Further, CTCF interacts with the large subunit of RNA polymerase II (RNA Pol II) for the activation of transcription in mammalian cells (24), and it has recently been shown that during the lytic infection of HSV-1, CTCF recruits RNA Pol II to the genome for transcription (31), further supporting the hypothesis that CTCF sites have differential functions during the lytic infection in epithelial cells and latent infection in neurons. We also showed that the CTRS3 site upstream from the ICP4 promoter had weak enhancer-blocking activity when the CTRS3 and LTE/CTRS3 constructs were compared in Vero cells, but when we tested this construct in Neuro 2a cells, we found no enhancer-blocking function, supporting our hypothesis that CTCF insulators in HSV-1 have dynamic lytic/latent functions. We hypothesize that during latency, the CTRS3 site functions as a silencer insulator to recruit corepressor protein complexes to repress ICP4 transcription. This hypothesis is supported by previous work showing enrichment of repressive histone marks on the ICP4 gene region as well as our own data showing that Suz12 is enriched on the CTRS3 site during latency and is evicted during reactivation.
We quantified the binding of CTCF to CTRL1 and CTRS1/2 upstream from the LAT promoter and downstream from the ICP4 region, respectively. We found that CTRL1 was significantly more enriched in CTCF than all other binding sites in HSV-1 during latency. Further, upon reactivation, the CTRL1 site did not completely evict CTCF, suggesting that this site retains some insulator function. While it is possible that CTCF is not globally evicted from the CTRL1 site because not all HSV-1 genomes are reactivating after NaB or explant treatment, we do not think that this is the case since in such an instance the other CTCF sites would also retain CTCF following reactivation. This is clearly not the case in the CTRS3 site, where CTCF binding is essentially abolished during reactivation. Further, our findings are consistent with CTCF regulation in other DNA viruses, where CTCF control of viral gene transcription has been described. Specifically, reports have shown that CTCF associates with several regions of Kaposi's sarcoma-associated herpesvirus (KSHV) in a cell cycle-dependent manner to repress lytic transcription (32, 33) and that the binding of CTCF is dynamic. Following lytic activation in KSHV, CTCF is not globally evicted; rather, it is gradually reduced at some sites but maintained at others. These observations in KSHV are similar to the CTCF binding and eviction dynamics we observed in HSV-1.
We hypothesized that the PRC2 complex was recruited to the genome and specifically to the CTCF binding motifs through a CTCF-Suz12 interaction to establish transcriptionally repressed domains around the IE genes as latency is established. CTCF insulators directly interact with the protein Suz12 in mammalian cells, and since the PRC2 is responsible for the trimethylation of H3K27 to silence transcription (27), CTCF recruitment of PRC2 through a Suz12 interaction in HSV-1 was consistent with the H3K27me3 status of lytic regions during latency. We found that Suz12 colocalized primarily to CTCF binding domains that flank the ICP0 and ICP4 gene regions of the genome. We speculate that Suz12 enrichment around the IE gene sites is a regulatory element maintaining repression of lytic transcription, and the mechanism by which CTCF establishes these domains is under investigation and will be the subject of a future manuscript. In contrast, Suz12 accumulation decreased at early times postreactivation, consistent with the loss of CTCF binding and changes to histone marks at those sites upon reactivation. These findings further support the idea that CTCF recruits the PRC2 through a Suz12 interaction to repress IE transcription during latency. We also note that SUZ12 was recruited to other sites that lie outside the CTCF binding domains, including the LAT promoter site. These data are consistent with previous findings by Cliffe et al., who reported that Suz12 occupied the LAT promoter region in mouse ganglia at 14 days postinfection (14). This finding suggests that other regulatory elements maintain the ability to recruit the PRC2. One possible explanation for our observations is that the LAT, a long noncoding RNA (lncRNA), interacts with the PRC2 in a manner similar to how host ncRNAs interact with and recruit the PRC2 for gene repression (34).
Using a mutant virus, 17ΔPst, we showed that CTCF recruitment on the genome is LAT dependent, further indicating that LAT plays a pivotal role in silencing lytic transcription through coordination of CTCF binding on the HSV-1 genome. While we hypothesize that LAT transcription is essential for CTCF recruitment, our data do not preclude that the deletion of the LAT promoter could result in changes to RNA Pol II binding or changes to protein recruitment that would alter CTCF recruitment and binding. Nevertheless, the finding that there is less CTCF bound to the mutant 17ΔPst virus CTCF binding sites is an interesting finding. If we consider that CTCF eviction is an important component of HSV-1 reactivation, then one might expect that viruses that do not reactivate have more CTCF bound to these sites to maintain repression of the genome. However, this is not the case, and we consistently see significantly less CTCF bound to viruses that do not reactivate in a site-specific manner (23). While our data show us that CTCF occupation is a LAT-driven phenomenon, we hypothesize that the ability of the wt virus to establish higher-order chromatin structures for the regulation of gene expression are also involved. CTCF can self-dimerize on insulators to form higher-order chromatin structures, known as chromatin loops, which bring distant enhancers and promoters into spatial proximity to regulate gene expression (35–37). Consequently, CTCF insulators regulate the local balance of active and repressive histone marks within chromatin loops through the recruitment of corepressors. The differential regulation of each CTCF binding motif in HSV-1 could serve to place the LAT enhancer into spatial proximity with lytic promoters to regulate gene expression, with LAT expression being a critical element to the establishment of functional chromatin loops in HSV-1. In support of our hypothesis, there is evidence that support roles for chromatin loops in the regulation of other herpesviruses such as KSHV and Epstein-Barr virus (EBV) (38, 39). In KSHV, CTCF and cohesion proteins colocalize to mediate DNA looping (33). Conventional chromosome conformation capture (3C) assays in cell culture have also identified a chromatin loop that likely mediates type I and type III latency in EBV (40). We speculate that the differential regulation of each CTCF site in HSV-1 may be related to genomic organization in 3D to regulate transcription in the HSV-1 genome. These ideas are currently being explored in the animal models of HSV-1.
Conclusion.
We found that CTCF binding sites in HSV-1 are independently and dynamically regulated in a cell-type-specific manner, a finding consistent with CTCF regulation of other DNA viruses. We explored the possibility that CTCF binding sites may recruit the coregulatory protein complex, polycomb-repressive complex 2 (PRC2), through a CTCF-Suz12 interaction to establish transcriptionally repressed domains during latency. Here, we found evidence that Suz12 colocalized to CTCF binding motifs flanking the IE regions of ICP0 and ICP4 in latent HSV-1. Our data indicate that CTCF recruitment of the PRC2 through a Suz12-CTCF interaction may be a mechanism by which lytic transcription is silenced during HSV-1 latency.
MATERIALS AND METHODS
Viruses and cells.
All experiments were performed using ganglia from mice infected with HSV-1 strains 17Syn+ and 17ΔPst (LAT promoter mutant from the 17Syn+ parent with a 202-bp deletion of nt 118664 to 118866; GenBank accession number NC_001806) (obtained from David C. Bloom, University of Florida) and the 17ΔPst rescued virus. All viruses were amplified and titrated on rabbit skin cells (CCL-68; ATCC) using Eagle's minimal essential medium (Life Technologies) supplemented with 5% calf serum, 250 U of penicillin/ml, 250 μg of streptomycin/ml, and 292 μg of l-glutamine/ml (Life Technologies).
Mouse ocular infections.
Female BALB/c mice 4 to 6 weeks of age (Taconic) were anesthetized using intramuscular injections of ketamine and xylazine (200 mg/kg and 10 mg/kg body weight, respectively). Using a 27-gauge needle, a light 2-by-2 crosshatch pattern was made on the corneal epithelium. Viral suspensions of 150,000 PFU/eye in a 5-μl volume were added to each eye for 17Syn+, 17ΔPst, and the 17ΔPst rescued virus. Animals were considered latent at >28 days postinfection (12). To verify that equivalent infections were established following infections with each virus, genome copies per ganglia were measured by quantitative real-time PCR using primers and a probe specific for HSV-1 DNA polymerase.
HSV-1 reactivation.
Viral reactivation was induced using either intraperitoneal injections of sodium butyrate (NaB) (41) or by using the explant method, as previously described (23). Following NaB treatment, mice were euthanized at 1 h, 2 h, 3 h, 4 h, or 6 h post-NaB treatment, and the trigeminal ganglia (TG) were rapidly removed and homogenized in cold phosphate-buffered saline (PBS) with protease inhibitors for ChIP procedures (see method below). Latent TG samples were not treated with NaB prior to homogenization for ChIP processing as 0-h samples. In explant-induced reactivation experiments, the TG were harvested from latently infected mice, placed in Dulbecco's modified Eagle's medium (DMEM) supplemented with 1% fetal bovine serum and 2% penicillin-streptomycin and placed in an incubator at 37°C with 5% CO2 for 1 h, 3 h, or 6 h. Following incubation, the TG were removed from the DMEM, placed in cold PBS with protease inhibitors, and processed for ChIP assays.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed as previously described for mice using the specific antibody anti-CTCF or anti-Suz12 (Millipore) (12). Each ChIP assay contained pooled TG (total of 6) of three mice. The TG were rapidly removed following euthanasia, the ganglia were homogenized, and the chromatin was cross-linked in 1% formaldehyde (Sigma-Aldrich). The cross-linked cell lysates were sonicated to shear the chromatin into fragments between 300 and 800 bp. Fragment size following sonication was confirmed by agarose gel electrophoresis using a 1.5% agarose gel. The sheared chromatin was precleared with salmon sperm DNA-protein A agarose beads (Millipore) prior to antibody incubation. An aliquot representing one-fifth of the total sample volume was removed as a sample input (I). The remaining sample was incubated in a cold room overnight with shaking with 2 μg of antibody per 1 ml of sample or with IgG as a nonspecific antibody binding control. The chromatin-antibody complexes were collected with salmon sperm DNA-protein A agarose beads and eluted to represent the bound (B) fraction. Both bound and input fractions were treated with 5 M NaCl, RNase A, and proteinase K, and the DNA was finally purified using a QiaQuick PCR purification kit (Qiagen). Prior to real-time PCR analyses of viral targets, all anti-CTCF ChIP assays were validated by determining the B/I ratios of the cellular controls Tsix imprinting/choice center CTCF site A (positive control) and MT498 (negative control) with the following primers: Tsix imprinting/choice center CTCF site A (GenBank accession number AJ421479) forward, CGCAGGGCAGCCAGAA, and reverse TCTGGTGTTATCCCTTCCTGTCTT, with probe CAGCCATTCACAATCC; mouse MT498 (NT_039554) forward, TACAAGATGCAAGTCTAGATATTTTAAGTCTATGTAT, and reverse, ACACACACACACACACACACA, with probe CACACACACACAAACAC; mouse adenine phosphoribosyltransferase (APRT) forward, CTCAAGAAATCTAACCCCTGACTCA, and reverse, GCGGGACAGGCTGAGA, with probe CCCCACACACACCTC (42). Only assays having a greater than 2-fold abundance of CTCF bound to the Tsix imprinting/choice center CTCF site A relative to the MT498 level were used in further analyses. The normalized B/I ratios for the viral gene gC was used as a negative viral control due to the fact that it lies ∼15 kb away from the nearest CTCF binding domain in HSV-1 and therefore should not be associated with CTCF occupation in ChIP analyses. For the Suz12 assays, all assays were validated with a negative-control ChIP using rabbit IgG (Abcam). All values for viral targets were normalized to the level of the negative control.
Real-time PCR analysis of ChIP experiments.
All real-time PCR experiments were performed using TaqMan universal PCR master mix, No AmpErase uracil N-glycosylase, on a qTower real-time PCR detection system (Analytik Jena) using custom-designed primer-probe mixes (Applied Biosystems) with the following general protocol: 95°C for 10 min followed by 45 cycles of 95°C for 15 s and then 60°C for 1 min. Threshold values used for PCR analyses were set within the linear range of PCR target amplification based on a standard curve generated for each plate. Primer and probe sequence used for each viral target are listed in Table 1.
TABLE 1.
Primers and probes used in this study
| Target and primer or probe | Primer sequence | Positiona (nt) |
|---|---|---|
| HSV-1 CTRL1 | 117004–117071 | |
| Forward | 5′-AGCCAAGTTAACGGGCTACG-3′ | |
| Reverse | 5′-ATGCCAGCCCAACAAAATC-3′ | |
| Probe | 5′-CCTTCGGGAATGGGACTGG-3′ | |
| HSV-1 CTRL2 | 120455–120643 | |
| Forward | 5′-CCGCGGCTCTGTGGTTA-3′ | |
| Reverse | 5′-GGATGCGTGGGAGTGGG-3′ | |
| Probe | 5′-ACACCAGAGCCTGCCCAACATGGCA-3′ | |
| HSV-1 CTa'm | 126368–126560 | |
| Forward | 5′-GCTGCCACAGGTGAAACC-3′ | |
| Reverse | 5′-TGTAGCAGGAGCGGTGTG-3′ | |
| Probe | 5′-ACCTGCCCAACAACACAACT-3′ | |
| HSV-1 CTRS1/2 | 127138–127303 | |
| Forward | 5′-GCCCCTCGAATAAACAACGCTA-3′ | |
| Reverse | 5′-GTTGTGGACTGGGAAGGCGC-3′ | |
| Probe | 5′-GGTTGTTGCCGTTTATTGCG-3′ | |
| HSV-1 CTRS3 | 132247–132385 | |
| Forward | 5′-ATCGCATCGGAAAGGGACACG-3′ | |
| Reverse | 5′-CCAAGGTGCTTACCCGTGCAAA-3′ | |
| Probe | 5′-ACAGAAACCCACCGGTCCGCCTTT-3′ | |
| HSV-1 gC | 96331–96643 | |
| Forward | 5′-CCTTGCCGTGGTCCTGTGGA-3′ | |
| Reverse | 5′-GGTGGTGTTGTTCTTGGGTTTG-3′ | |
| Probe | 5′-CCCCACGTCCACCCCCGACC-3′ | |
| HSV-1 DNA pol | 65880–65953 | |
| Forward | 5′-AGAGGGACATCCAGGACTTTGT-3′ | |
| Reverse | 5′-CAGGCGCTTGTTGGTGTAC-3′ | |
| Probe | 5′-ACCGCCGAACTGAGCA-3′ | |
| HSV-1 LAT Intron | 119721–119795 | |
| Forward | 5′-ACCCACGTACTCCAAGAAGGC-3′ | |
| Reverse | 5′-TAAGACCCAAGCATAGAGAGCCA-3′ | |
| Probe | 5′-TCCCACCCCGCCTGTGTTTTT-3′ |
Nucleotide positions are based on GenBank accession number NC_001806.
Determination of bound/input ratios from ChIP.
ChIP data are presented as a ratio of the relative copy numbers of the PCR target in two fractions (bound and input), where the bound fractions are representative of aliquots incubated overnight with the antibody or IgG and the input fractions are precleared chromatin only (no specific antibody incubation). Standard curves were generated for each PCR run for each gene region analyzed using serial dilutions of purified HSV-1 DNA or purified mouse DNA using primer/probes specific for that region. Each ChIP assay was analyzed as follows. First, duplicate PCRs were performed using DNA purified from either the bound (B) or input (I) fraction as the target. The average cycle threshold (CT) for the bound fraction and the average CT for the input fraction were then used to determine the relative quantity of target DNA in each fraction by using the equation for the standard curve specific to the primer and probe set used. The quantity was expressed as a ratio of the relative bound quantity to the relative input quantity (B/I ratio). All relative B/I ratios from each gene region were normalized to the B/I ratio for the cellular control Tsix site A for the anti-CTCF incubations or the B/I ratio for IgG for the anti-Suz12 incubations.
qPCR for genome copies/ganglia.
Real-time primer probe sets specific to HSV-1 DNA polymerase were used on all input fractions from ChIP samples to determine equivalent infections during latency and following HSV-1 reactivation stimuli through 6 h. Real-time PCR was performed as described above. The relative HSV-1 copy numbers for each sample were further normalized to the relative copy numbers of the host control APRT.
Quantitative reverse transcription-PCR (qRT-PCR) analysis.
Mouse TG were isolated and placed in RNAlater and stored according to the manufacturer's specifications. RNA was extracted by removing RNAlater from samples and adding TRIzol reagent (Sigma-Aldrich) to each sample. Briefly, each TG was homogenized in 1.2 ml of TRIzol, and following the addition of 0.2 volume of chloroform, samples were centrifuged for phase separation. RNA was precipitated from the aqueous phase using 0.7 volume of isopropanol, followed by DNase treatment using DNA-free (Ambion), according to the manufacturer's directions. Reverse transcription using random primers was performed with a high-capacity cDNA reverse transcription kit (ABI), according to the manufacturer's instructions. Briefly, 20-μl reaction mixtures contained DNase-treated RNA, manufacturer-supplied buffer, deoxynucleoside triphosphate mix, 1 μM random hexamer primer, 1 U of RNase inhibitor (Ambion), and Multiscribe reverse transcriptase. Real-time PCRs were performed on cDNA according to the above-described procedures and protocols using the LAT intron primers and probes shown in Table 1 (mouse APRT forward, 5′-CTCAAGAAATCTAACCCCTGACTCA-3′; reverse, 5′-GCGGGACAGGCTGAGA-3′; probe, 5′-CCCCACACACACCTC-3′).
PCR through CTCF motifs.
Endpoint PCR was done using viral DNA purified from either wt or mutant virus to determine if there was a difference in the sizes of the consensus motifs as a result of the deletion of the LAT promoter sequence. Endpoint PCR was done using a HotStar HiFidelity polymerase kit (Qiagen). Primer sequences and PCR conditions for each fragment amplified by PCR are listed in Table 2.
TABLE 2.
PCR primers for HSV-1 targets
| DNA target | Primer direction | Primer sequence | Position (nt) | PCR conditions |
|---|---|---|---|---|
| CTRL1 | Forward | 5′-TAGGTCCTTGGGGTAATGGAAAAGG-3′ | 116959–117773 | 95°C for 5 min; 40 cycles of 98°C for 1 min, 60°C for 2 min, and 72°C for 2 min; 72°C for 12 min |
| Reverse | 5′-AAAACGAGGAGGAGGAGGAGAAGG-3′ | |||
| CTRL2 | Forward | 5′-CAACACCCACGTCTGTGGTGT-3′ | 120210–120941 | 95°C for 5 min; 30 cycles of 98°C for 10 s, 65°C for 1 min, and 72°C for 2 min; 72°C for 12 min |
| Reverse | 5′-GTCTCGGCACCCGTGGTC-3′ | |||
| CTRS1/2 | Forward | 5′-CAGGTGAAACCAACAGAGCA-3′ | 126376–126750 | 95°C for 5 min; 35 cycles of 94°C for 1 min, 60°C for 2 min, and 72°C for 2 min; 72°C 12 min |
| Reverse | 5′-GCCGCCAATCTCAGGTCAGA-3′ | |||
| CTRS3 | Forward | 5′-GTCCCCGCCCTCCTCCGTC-3′ | 131000–132600 | 95°C for 5 min; 35 cycles of 98°C for 10 s, 65°C for 1.5 min, and 72°C for 1 min; 72°C for 12 min |
| Reverse | 5′-ACGAGCAGGAAGCGGTCCACG-3′ |
Plasmid constructs and enhancer blocking assays.
The HSV-1 LAT enhancer (nucleotides [nt] 118888 to 119477) was directionally cloned into the 5′ KpnI and 3′ MluI polylinker sites of the pGL3 control vector (Promega) using PCR-generated KpnI and MluI linkers to generate the pGL3-control/LTE enhancer plasmid, as previously described (22). Additionally, a 2.1-kb fragment (nt 118888 to 120942) with the LAT enhancer and CTRL2 insulator in their native HSV-1 configurations was directionally cloned into the 5′ KpnI and 3′ XhoI polylinker sites of the pGL3 control plasmid using PCR-generated KpnI and XhoI linkers to create pGL3-control/LTE+CTRL2 (23). Other constructs generated included the CTRL1 and the CTRS1 regions of HSV-1. First, a 932-bp fragment (nt 117056 to 117988) containing sequences flanking the core CTCF binding cluster of CTRL1 (Fig. 1A) was generated in the pGL3 control plasmid and sequence verified (Biomatik). The pGL3-control/CTRL1 plasmid was then used to generate pGL3-control/LTE+CTRL1 by directionally cloning the LTE fragment into the 5′ KpnI and 3′ SacI sites of the pGL3-control/CTRL1 plasmid using PCR-generated KpnI and SacI linkers. Another plasmid containing sequences flanking the core CTCF binding motif CTRS1 (Fig. 1A) was generated by PCR with NheI and Xho1 linkers (nt 126376 to 126758). The pGL3-control/CTRS1 and pGL3-control/LTE+CTRS1 plasmids were generated by directionally cloning the CTRS1 into the 5′ NheI and 3′ XhoI sites using PCR-generated NheI and XhoI linkers to create the indicated plasmids. The CTa′m and the CTRS3 constructs were generated as follows: a 1.3-kb fragment (nt 125013 to 126375) containing sequences flanking the 217-bp core CTCF binding cluster of CTa′m (Fig. 1A) was generated by PCR with NheI and Xho1 linkers. The pGL3-control/CTa′m and pGL3-control/LTE+CTa′m plasmids were generated by directionally cloning the CTa′m fragment into the 5′ NheI and 3′ XhoI sites of the pGL3 control or the pGL3-control/LTE enhancer, respectively, using PCR-generated NheI and XhoI linkers. A 1.6-kb fragment (nt 131000 to 132600) containing sequences flanking the core 111-bp cluster of the CTCF binding motif CTRS3 (Fig. 1A) was generated by PCR with NheI and Xho1 linkers. The pGL3-control/CTRS3 and pGL3-control/LTE+CTRS3 plasmids were generated by directionally cloning the CTRS3 site into the 5′ NheI and 3′ XhoI sites using PCR-generated NheI and XhoI linkers. Transient transfections were done using 1 μg of each luciferase test construct described above in 24-well dishes seeded 24 h earlier with either Vero cells or Neuro 2a cells (ATCC) at 2 × 105 cells/ml (ATCC). Five hundred nanograms of phRL-TK Renilla construct was cotransfected as a control for transfection efficiency. Each luciferase construct was tested in triplicate and repeated five times. Transfections were carried out using 5 μl of SuperFect transfection reagent (Qiagen) per reaction well according to the manufacturer's protocols. Following a 24-h incubation, the cells were lysed, and luciferase levels were tested using a Dual Luciferase Reporter Assay kit (Promega) according to the manufacturer's protocols, and the levels were measured using a Berthold Technologies TriStar LB 941 Luminometer with MikroWin 2000.
Statistical analysis.
One-way analysis of variance (ANOVA) was performed using SigmaPlot, version 12.5, by analyzing the individual normalized B/I ratios for a specific gene region during latency compared to the normalized B/I ratio for that same gene region following NaB treatment (or explant-induced reactivation) at each individual time point analyzed by ChIP. The P values are indicated on the graphs above the time point at which a significant change was determined by one-way ANOVA. For luciferase assays, unpaired one-tailed Student's t tests in pairwise comparisons of the CTCF construct and CTCF-LTE constructs were done.
Ethics statement.
The animal studies were approved by the Institutional Animal Care and Use Committee of the Louisiana State University Health Sciences Center, New Orleans (institutional animal welfare assurance no. A3094-01).
ACKNOWLEDGMENTS
This work was supported in part by grant R56 AI101174 from the NIH-NIAID, COBRE P30 GM106392, and an unrestricted grant from Research to Prevent Blindness and the Department of Pharmacology, LSUHSC New Orleans.
REFERENCES
- 1.Bloom DC. 2016. Alphaherpesvirus latency: a dynamic state of transcription and reactivation. Adv Virus Res 94:53–80. doi: 10.1016/bs.aivir.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Austin A, Lietman T, Rose-Nussbaumer J. 2017. Update on the management of infectious keratitis. Ophthalmology 124:1678–1689. doi: 10.1016/j.ophtha.2017.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibeault RL, Conn KL, Bildersheim MD, Schang LM. 2016. An essential viral transcription activator modulates chromatin dynamics. PLoS Pathog 12:e1005842. doi: 10.1371/journal.ppat.1005842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stevens JG, Wagner EK, Devi Rao GB, Cook ML, Feldman LT. 1987. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science 235:1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 5.Jarman RG, Loutsch JM, Devi-Rao GB, Marquart ME, Banaszak MP, Zheng X, Hill JM, Wagner EK, Bloom DC. 2002. The region of the HSV-1 latency-associated transcript required for epinephrine-induced reactivation in the rabbit does not include the 2.0-kb intron. Virology 292:59–69. doi: 10.1006/viro.2001.1265. [DOI] [PubMed] [Google Scholar]
- 6.Perng GC, Ghiasi H, Slanina SM, Nesburn AB, Wechsler SL. 1996. The spontaneous reactivation function of the herpes simplex virus type 1 LAT gene resides completely within the first 1.5 kilobases of the 8.3-kilobase primary transcript. J Virol 70:976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perng GC, Dunkel EC, Geary PA, Slanina SM, Ghiasi H, Kaiwar R, Nesburn AB, Wechsler SL. 1994. The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. J Virol 68:8045–8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwiatkowski DL, Thompson HW, Bloom DC. 2009. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J Virol 83:8173–8181. doi: 10.1128/JVI.00686-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cliffe AR, Garber DA, Knipe DM. 2009. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J Virol 83:8182–8190. doi: 10.1128/JVI.00712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang QY, Zhou C, Johnson KE, Colgrove RC, Coen DM, Knipe DM. 2005. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc Natl Acad Sci U S A 102:16055–16059. doi: 10.1073/pnas.0505850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kubat NJ, Tran RK, McAnany P, Bloom DC. 2004. Specific histone tail modification and not DNA methylation is a determinant of HSV-1 latent gene expression. J Virol 78:1139–1149. doi: 10.1128/JVI.78.3.1139-1149.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neumann DM, Bhattacharjee PS, Giordani NV, Bloom DC, Hill JM. 2007. In vivo changes in the patterns of chromatin structure associated with the latent herpes simplex virus type 1 genome in mouse trigeminal ganglia can be detected at early times after butyrate treatment. J Virol 81:13248–13253. doi: 10.1128/JVI.01569-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubat NJ, Amelio AL, Giordani NV, Bloom DC. 2004. The herpes simplex virus type 1 latency-associated transcript (LAT) enhancer/rcr is hyperacetylated during latency independently of LAT transcription. J Virol 78:12508–12518. doi: 10.1128/JVI.78.22.12508-12518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cliffe AR, Coen DM, Knipe DM. 2013. Kinetics of facultative heterochromatin and polycomb group protein association with the herpes simplex viral genome during establishment of latent infection. mBio 4:e00590-12. doi: 10.1128/mBio.00590-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knipe DM, Cliffe A. 2008. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol 6:211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- 16.Cliffe AR, Knipe DM. 2008. Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J Virol 82:12030–12038. doi: 10.1128/JVI.01575-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kristie TM. 2016. Chromatin modulation of herpesvirus lytic gene expression managing nucleosome density and heterochromatic histone modifications. mBio 7:e00098-. doi: 10.1128/mBio.00098-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kristie TM, Liang Y, Vogel JL. 2010. Control of alpha-herpesvirus IE gene expression by HCF-1 coupled chromatin modification activities. Biochim Biophys Acta 1799:257–265. doi: 10.1016/j.bbagrm.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen SH, Kramer MF, Schaffer PA, Coen DM. 1997. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J Virol 71:5878–5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.West AG, Gaszner M, Felsenfeld G. 2002. Insulators: many functions, many mechanisms. Genes Dev 16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- 21.Ghirlando R, Felsenfeld G. 2016. CTCF: making the right connections. Genes Dev 30:881–891. doi: 10.1101/gad.277863.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amelio AL, McAnany PK, Bloom DC. 2006. A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities. J Virol 80:2358–2368. doi: 10.1128/JVI.80.5.2358-2368.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ertel MK, Cammarata AL, Hron RJ, Neumann DM. 2012. CTCF occupation of the HSV-1 genome is disrupted at early times post-reactivation in a transcription-dependent manner. J Virol 86:12741–12759. doi: 10.1128/JVI.01655-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chernukhin I, Shamsuddin S, Kang SY, Bergstrom R, Kwon YW, Yu W, Whitehead J, Mukhopadhyay R, Docquier F, Farrar D, Morrison I, Vigneron M, Wu SY, Chiang CM, Loukinov D, Lobanenkov V, Ohlsson R, Klenova E. 2007. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol Cell Biol 27:1631–1648. doi: 10.1128/MCB.01993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lutz M, Burke LJ, Barreto G, Goeman F, Greb H, Arnold R, Schultheiss H, Brehm A, Kouzarides T, Lobanenkov V, Renkawitz R. 2000. Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res 28:1707–1713. doi: 10.1093/nar/28.8.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li T, Hu JF, Qiu X, Ling J, Chen H, Wang S, Hou A, Vu TH, Hoffman AR. 2008. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol Cell Biol 28:6473–6482. doi: 10.1128/MCB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simon JA, Kingston RE. 2009. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 28.Amelio AL, Giordani NV, Kubat NJ, O'Neil J, Bloom DC. 2006. Deacetylation of the herpes simplex virus type 1 latency-associated transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times postexplant. J Virol 80:2063–2068. doi: 10.1128/JVI.80.4.2063-2068.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devi-Rao GB, Bloom DC, Stevens JG, Wagner EK. 1994. Herpes simplex virus type 1 DNA replication and gene expression during explant-induced reactivation of latently infected murine sensory ganglia. J Virol 68:1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevens HC, Fiskerstrand C, Bubb VJ, Dalziel R, Quinn JP. 2009. A regulatory domain spanning the repeat sequence RE1 from herpes simplex virus type 1 has cell specific differential functions in trigeminal neurons and fibroblasts. FEBS Lett 583:3335–3338. doi: 10.1016/j.febslet.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lang F, Li X, Vladimirova O, Hu B, Chen G, Xiao Y, Singh V, Lu D, Li L, Han H, Wickramasinghe JM, Smith ST, Zheng C, Li Q, Lieberman PM, Fraser NW, Zhou J. 2017. CTCF interacts with the lytic HSV-1 genome to promote viral transcription. Sci Rep 7:39861. doi: 10.1038/srep39861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang H, Lieberman PM. 2009. Cell cycle control of Kaposi's sarcoma-associated herpesvirus latency transcription by CTCF-cohesin interactions. J Virol 83:6199–6210. doi: 10.1128/JVI.00052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. 2008. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J 27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davidovich C, Cech TR. 2015. The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA 21:2007–2022. doi: 10.1261/rna.053918.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohlsson R, Renkawitz R, Lobanenkov V. 2001. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet 17:520–527. doi: 10.1016/S0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 36.Ohlsson R, Bartkuhn M, Renkawitz R. 2010. CTCF shapes chromatin by multiple mechanisms: the impact of 20 years of CTCF research on understanding the workings of chromatin. Chromosoma 119:351–360. doi: 10.1007/s00412-010-0262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim YW, Kim A. 2013. Histone acetylation contributes to chromatin looping between the locus control region and globin gene by influencing hypersensitive site formation. Biochim Biophys Acta 1829:963–969. doi: 10.1016/j.bbagrm.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 38.Chen HS, Martin KA, Lu F, Lupey LN, Mueller JM, Lieberman PM, Tempera I. 2014. Epigenetic deregulation of the LMP1/LMP2 locus of Epstein-Barr virus by mutation of a single CTCF-cohesin binding site. J Virol 88:1703–1713. doi: 10.1128/JVI.02209-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tempera I, Lieberman PM. 2010. Chromatin organization of gammaherpesvirus latent genomes. Biochim Biophys Acta 1799:236–245. doi: 10.1016/j.bbagrm.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tempera I, Wiedmer A, Dheekollu J, Lieberman PM. 2010. CTCF prevents the epigenetic drift of EBV latency promoter Qp. PLoS Pathog 6:e1001048. doi: 10.1371/journal.ppat.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neumann DM, Bhattacharjee PS, Hill JM. 2007. Sodium butyrate: a chemical inducer of in vivo reactivation of herpes simplex virus type 1 in the ocular mouse model. J Virol 81:6106–6110. doi: 10.1128/JVI.00070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chao W, Huynh KD, Spencer RJ, Davidow LS, Lee JT. 2002. CTCF, a candidate trans-acting factor for X-inactivation choice. Science 295:345–347. doi: 10.1126/science.1065982. [DOI] [PubMed] [Google Scholar]