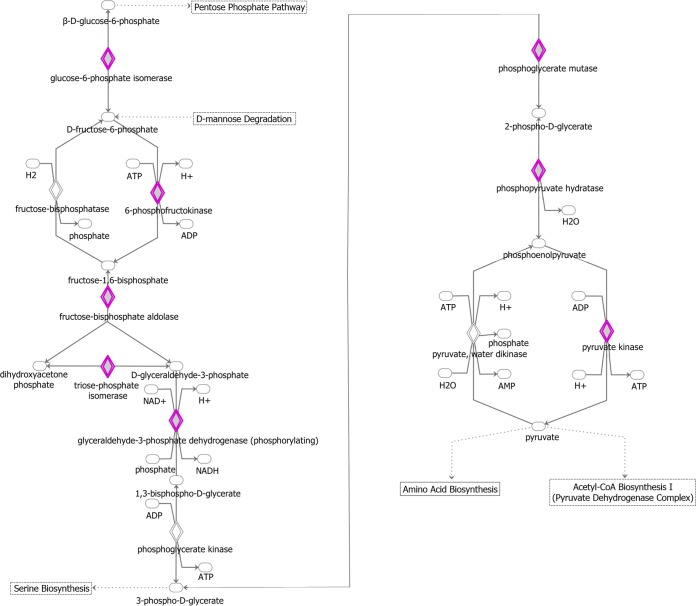
FIG 11.
Ingenuity Pathway Analysis diagram of the glycolysis pathway. IPA software was used to identify pathways that are enriched in the set of 1,433 genes that were present in three of three biological replicates on WT-infected TIVE-EX-LTC cells. Genes that are targeted by KSHV miRNAs are highlighted in purple. Ovals, transcription regulators; downward-pointing triangles, kinases; upward-pointing triangles, phosphatases; dotted squares, growth factors; circles, other; double-walled shapes, complexes/groups; diamonds, enzymes.