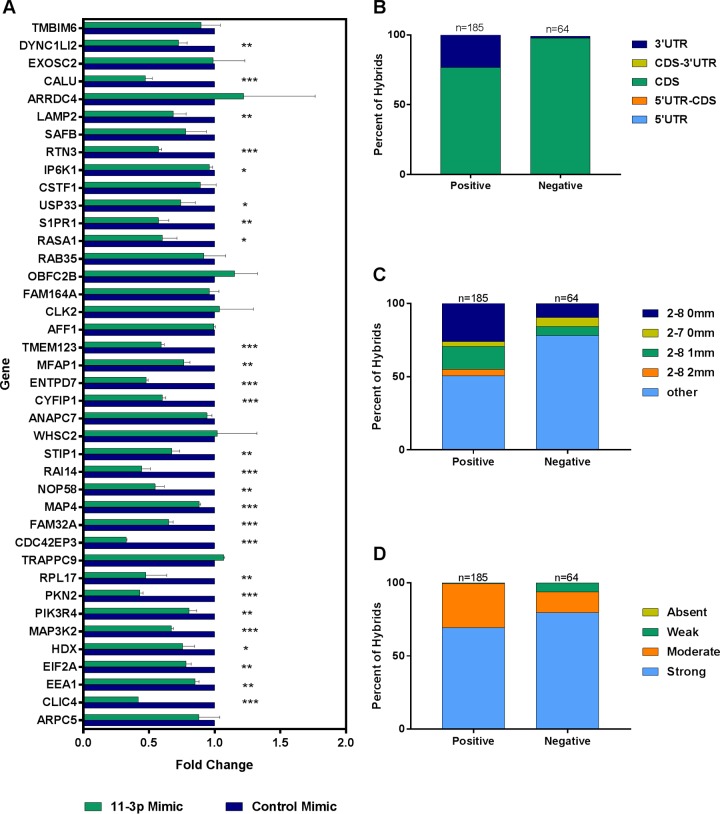
FIG 8.
Repression of selected mRNA targets validated by RT-qPCR. TIVE-EX-LTC cells infected with ΔmiR-K12-11 were transfected with either a KSHV miR-K12-11-3p mimic or a nontargeting control mimic. RNA was prepared from the cells, and expression levels of specific mRNA targets of miR-K12-11 discovered by qCLASH were measured by RT-qPCR. (A) Fold changes in expression levels of the indicated mRNAs in miR-K12-11-3p mimic-transfected cells compared to control mimic-transfected cells. Data shown are the averages of results from two biological replicates. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (B and C) Genes that were positive for repression in the presence of the miR-K12-11-3p mimic were compared to those that were negative for repression. (B) Percentages of hybrids that contain an mRNA fragment originating from the CDS or the 3′ UTR. (C) Percentages of hybrids exhibiting the different types of indicated seed matches (2-8 0 mm, nucleotides 2 to 8 with no mismatches; 2-7 0 mm, nucleotides 2 to 7 with no mismatches; 2-8 1 mm, nucleotides 2 to 8 with 1 mismatch; 2-8 2 mm nucleotides 2 to 8, with 2 mismatches). (D) Comparison of genes that were positive for repression versus those that were negative based on binding strength at the 3′ end of the hybrid miRNA. Strong, >8 bound nucleotides; moderate, 5 to 8 bound nucleotides; weak, 1 to 4 bound nucleotides; absent, 0 bound nucleotides.