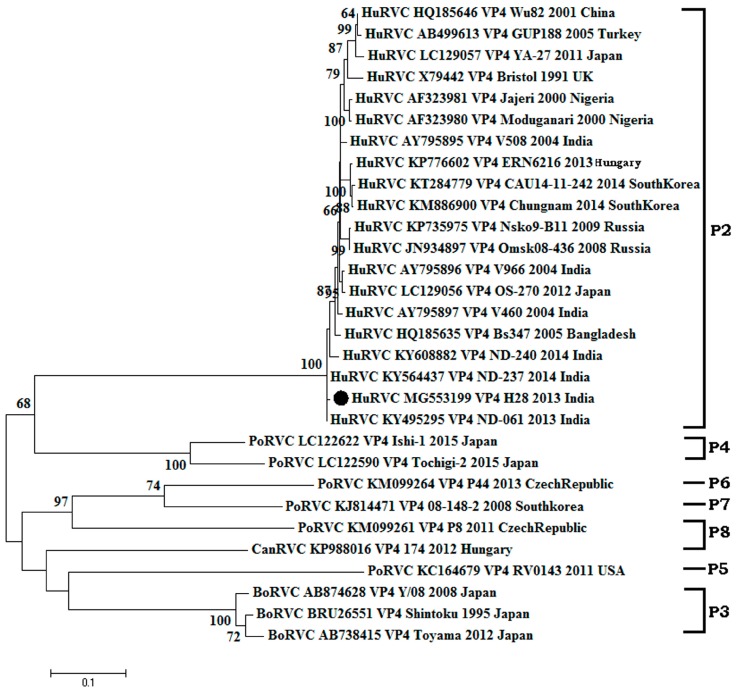
Figure 4.
Phylogenetic analysis of rotavirus C based on partial length (1224 bp) VP4 genes of human, swine, cattle and dog origin at the nucleotide level. The Tamura-3+G+I algorithm was identified using the find best DNA/protein model tool available in MEGA 6, which was confirmed with the FindModel online tool [57]. Numbers on branches indicate the percentages of bootstrap support from 1000 replicates. VP6 gene-based I typing of RVC is denoted along with clusters. Host species depicted are: human (Hu); cattle (Bo); pig (Por); dog (Can). Strains/isolates are represented according to their host species, accession number, gene, strain, year of isolation and country of origin. The isolate of the current study is denoted by the solid dot. Genotype designations are shown on the right. The scale bar indicates nucleotide substitutions per site.