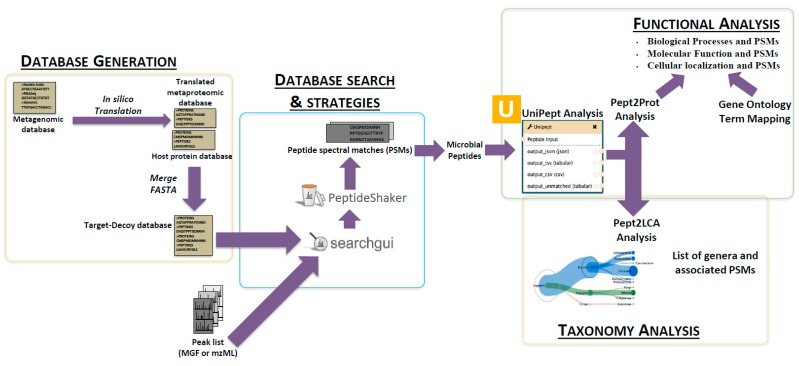
Figure 1.
Generalized metaproteomics schema: Identification of metaproteome peptides is a complex workflow consisting of metaproteome sequence database generation (in FAST-ALL (FASTA) format) and peak processing of tandem mass spectrometry (MS/MS) data (in Mascot Generic Format (MGF) of mzML format). These two output files are used to match observed MS/MS spectra to predicted peptide sequences. This generates a list of bacterial peptide–spectral matches (PSMs). Later, the bacterial PSMs can be parsed out and subjected to functional analysis and taxonomic analysis for biological insight.