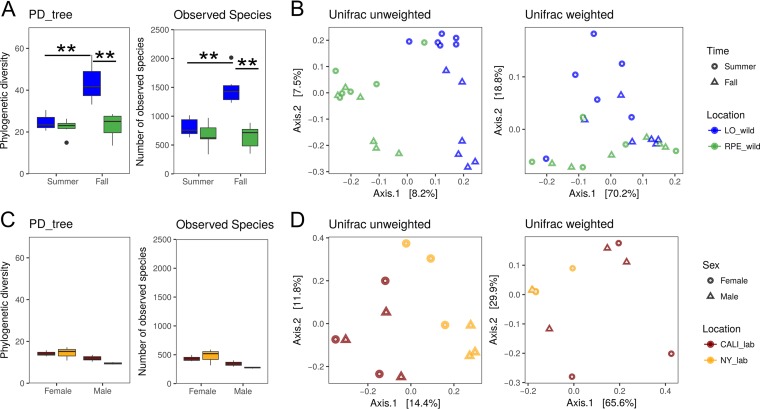
FIG 1 .
Bacterial community variation among wild and laboratory D. suzukii flies. (A) Box plots of phylogenetic diversity (PD_tree) and number of observed species of microbiota from wild D. suzukii flies. (For PD data, location P = 0.0006, time P < 0.0001, and sex P = 0.0050; for observed-species data, location P = 0.0002, time P = <0.0001, and sex P = 0.0103). (B) Unweighted and weighted UniFrac-based principal-component analysis (PCoA) plots of bacterial communities in wild D. suzukii flies. (For unweighted data, location P = 0.001, time P = 0.001, and sex P = 0.149; for weighted data, location P = 0.095, time P = 0.03, and sex P = 0.196). (C) Box plots of phylogenetic diversity and number of observed species of microbiota from laboratory D. suzukii flies. (For PD data, location P = 0.0578 and sex P = 0.0128; for observed-species data, location P = 0.188 and sex P = 0.014). (D) Unweighted and weighted UniFrac-based PCoA plots of bacterial communities in laboratory D. suzukii flies. (For unweighted data, location P = 0.005 and sex P = 0.128; for weighted data, location P = 0.015 and sex P = 0.321). Statistically significant differences of alpha diversities were analyzed using three-way ANOVA with Tukey post hoc tests. Statistically significant differences in beta diversities were analyzed using Adonis analysis with 999 Monte Carlo permutations. **, P < 0.01.