Abstract
Background
Glycolate is a valuable chemical with extensive applications in many different fields. The traditional methods to synthesize glycolate are quite expensive and toxic. So, the biotechnological production of glycolate from sustainable feedstocks is of interest for its potential economic and environmental advantages. d-Xylose is the second most abundant sugar in nature and accounts for 18–30% of sugar in lignocellulose. New routes for the conversion of xylose to glycolate were explored.
Results
Overexpression of aceA and ghrA and deletion of aceB in Escherichia coli were examined for glycolate production from xylose, but the conversion was initially ineffective. Then, a new route for glycolate production was established in E. coli by introducing NAD+-dependent xylose dehydrogenase (xdh) and xylonolactonase (xylC) from Caulobacter crescentus. The constructed engineered strain Q2562 produced 28.82 ± 0.56 g/L glycolate from xylose with 0.60 ± 0.01 g/L/h productivity and 0.38 ± 0.07 g/g xylose yield. However, 27.18 ± 2.13 g/L acetate was accumulated after fermentation. Deletions of iclR and ackA were used to overcome the acetate excretion. An ackA knockout resulted in about 66% decrease in acetate formation. The final engineered strain Q2742 produced 43.60 ± 1.22 g/L glycolate, with 0.91 ± 0.02 g/L/h productivity and 0.46 ± 0.03 g/g xylose yield.
Conclusions
A new route for glycolate production from xylose was established, and an engineered strain Q2742 was constructed from this new explored pathway. The engineering strain showed the highest reported productivity of glycolate to date. This research opened up a new prospect for bio-refinery of xylose and an alternative choice for industrial production of glycolate.
Keywords: Xylose, Glycolate, Glycolaldehyde, Glyoxylate, Escherichia coli
Background
Glycolate is a notable α-hydroxy acid containing both alcohol and carboxyl groups [1]. Its properties make glycolate ideal for a broad spectrum of applications. It can be used as skincare product in the cosmetic industry, as rinsing agents in dyeing and tanning industry and as precursors in biopolymers synthesis [2–4]. The polymer of glycolate is an excellent packaging material due to the properties of gas-barrier and mechanical strength [5]. Glycolate is also used to be polymerized with lactic acid for medical applications [6]. With increasing demands, the glycolate market is expected to reach USD 277.8 million in 2020 (http://www.grandviewresearch.com/press-release/global-glycolic-acid-market).
Currently, glycolate is manufactured through two traditional methods. One is produced by chemical carbonylation of formaldehyde under high-temperature, high-pressure condition [7]. The other is produced from ethylene glycol by enzyme-catalyzed oxidation reaction [4], or from glycolonitrile by hydrolyzation reaction [8]. However, precursors of all the above reactions are quite expensive or toxic.
The biotechnological production of glycolate from sustainable feedstock, such as lignocellulose would be more economically feasible and environmental friendly [9–11]. d-glucose and d-xylose are the most abundant constituents of lignocellulose, which are principally derived from cellulose and hemicellulose, respectively [12]. d-glucose was easily metabolized by most microorganisms, and for this reason, metabolic engineering for glycolate production was mainly concentrated on the glyoxylate bypass with glucose as the carbon source [13–15]. However, xylose is the second most abundant sugar in nature, and accounts for 18–30% of sugar in lignocellulose [16]. Therefore, the capability to efficiently metabolize xylose is a desirable attribute of microorganisms for optimizing the economics of lignocellulose-based bio-refinery processes. In fact, only a small fraction of microorganisms has the native metabolic pathway of xylose [17]. d-xylose is initially converted to d-xylulose by the catalysis of different enzymes in microorganisms. Bacteria like Escherichia coli generally employ a one-step pathway using xylose isomerase (XI) enzyme, whereas yeasts and mycelial fungi need two enzymatic–catalytic steps by reduction reaction of d-xylose reductase (XR) and oxidation reaction of xylitol dehydrogenase (XDH) [18, 19]. d-xylulose is further metabolized to d-xylulose-5-phosphate and then directed to pentose phosphate pathway (PPP) in both bacteria and fungi [20]. Recently, researchers reported a new metabolism pathway of xylose through yielding α-ketoglutarate in the oligotrophic freshwater bacterium Caulobacter crescentus [21]. This novel pathway was initiated by the enzymes of NAD+-dependent xylose dehydrogenase (Xdh) and xylonolactonase (XylC) [21, 22]. All enzymes of this xylose degradation pathway in C. crescentus were encoded by the xyl operon and expressed by xylose induction [23, 24].
Escherichia coli uses the pentose phosphate pathway for the metabolism of xylose. An important intermediate acetyl-CoA is produced by the pentose phosphate pathway and the glycolysis. Then it is metabolized by the tricarboxylic acid (TCA) cycle (Fig. 1).
Fig. 1.
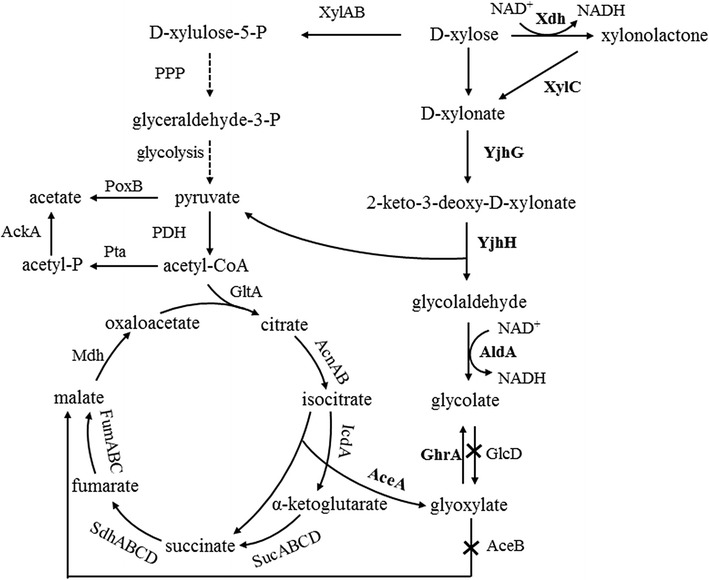
The metabolic pathway for production of glycolate from xylose in glycolaldehyde oxidation pathway and glyoxylate reduction pathway. Enzymes associated with the reactions were showed and the over expressions of which were marked in bold. d-xylose was converted to d-xylonate by xylose dehydrogenase (xdh) and xylonolactonase (xylC) from C. crescentus. All other genes were from the native E. coli BL21(DE3)
The intermediate isocitrate is not only converted to succinate by isocitrate dehydrogenase (icd) and α-ketoglutarate dehydrogenase (suc) in TCA cycle, but also is directly split into succinate and glyoxylate by isocitrate lyase (aceA) in glyoxylate bypass without CO2 emission. Finally, glyoxylate is reduced to glycolate by the bifunctional glyoxylate/hydroxypyruvate reductase (ghrA). The biosynthesis of glycolate in some previous studies was concentrated on glyoxylate bypass from glucose [13–15]. The previous results demonstrated that increased expression of aceA and ghrA and decreased expression of aceB showed positive effects on glycolate production with glucose as the carbon. E. coli have the native xylose assimilation pathway, and glycolate may also be produced from xylose via reduction of the intermediate glyoxylate, which is provided by the glyoxylate bypass. In order to verify this hypothesis, the glyoxylate reduction pathway was constructed and determined for the production of glycolate with xylose as the carbon. Besides, xylose can be oxidized to xylonate with generating one NADH in xylose assimilation pathway of C. crescentus. The latter is directly metabolized to pyruvate and glycolaldehyde via the dehydration and aldolytic cleavage (Fig. 1). Compared with the E. coli native XI pathway, this route shortens the xylose assimilation process which may improve the efficiency theoretically. So, a novel route was also designed for glycolate production in E. coli by introducing the xylose assimilation pathway of C. crescentus. Glycolate was gained from oxidation of the intermediate glycolaldehyde in this route, named the glycolaldehyde oxidation pathway. Meanwhile, the glyoxylate reduction pathway and the glycolaldehyde oxidation pathway were engineered simultaneously for determining and discussing the effects of the combination on glycolate production. This research may opened up a new prospect for bio-refining of xylose and improve the large scale production of glycolate.
Results and discussion
Production of glycolate from glyoxylate reduction pathway
Escherichia coli strains have the native xylose metabolism pathway. d-xylose was metabolized to glyoxylate catalyzed by a series of enzymes and further reduced to glycolate by GhrA. The glyoxylate reduction pathway for glycolate production was constructed. The coding regions of E. coli native genes aceA and ghrA were cloned to pACYCDuet1 and pETDuet1, respectively. The two recombinant plasmids pACYCDuet1-aceA (pLMa1) and pETDuet1-ghrA (pLMe1) were transformed to E. coli BL21(DE3), creating the engineered strain Q2728. The proteins of AceA and GhrA showed proper expressions as compared with the control strain E. coli BL21(DE3) harboring the empty plasmids pACYCDuet1 and pETDuet1 by gel electrophoresis analysis. The engineered strain Q2728 and wild-type E. coli BL21(DE3) were cultured with 15 g/L xylose as the sole carbon source. After 24 h, glycolate was not detected in E. coli BL21(DE3), suggesting that cells could not accumulate glycolate via the unmodified glyoxylate bypass (data was not shown). The engineered strain Q2728 accumulated 0.28 ± 0.01 g/L glycolate at a yield of 0.04 ± 0.01 g/g xylose (Table 1). In order to block the flux from glyoxylate to malate in glyoxylate bypass, the malate synthase (aceB) gene was knocked out to generate aceB mutant strain Q2589. The above recombinant plasmids pLMe1 and pLMa1 were transformed to Q2589, creating the engineered strain Q2705. As shown in Table 1, the glycolate production of Q2705 was improved to 0.51 ± 0.11 g/L, showing 1.8 times higher than that of Q2728. However, the xylose consumption of Q2705 also showed about 1.5 times higher as compared to Q2728. Through calculation, the glycolate yield of Q2705 was 0.05 ± 0.03 g/g xylose, similar to the value of Q2728. So, knockout of aceB had no significant improvement of the glycolate yield.
Table 1.
Production of glycolate after 24 h of IPTG induction under Erlenmeyer flasks cultivation on minimal medium with 15 g/L d-xylose
| Strains | Cell biomass (gCDW/L) | Cell yield (g/g xylose) | Glycolate concentration (g/L) | Glycolate yield (g/g xylose) | Acetate concentration (g/L) | Acetate yield (g/g xylose) |
|---|---|---|---|---|---|---|
| Q2728 | 1.05 ± 0.01 | 0.16 ± 0.07 | 0.28 ± 0.01 | 0.04 ± 0.01 | 0.66 ± 0.01 | 0.10 ± 0.02 |
| Q2705 | 1.12 ± 0.04 | 0.14 ± 0.02 | 0.51 ± 0.11 | 0.05 ± 0.03 | 1.33 ± 0.15 | 0.16 ± 0.05 |
| Q2562 | 1.03 ± 0.16 | 0.18 ± 0.09 | 2.53 ± 0.08 | 0.44 ± 0.10 | 0.95 ± 0.18 | 0.17 ± 0.04 |
| Q2590 | 1.09 ± 0.13 | 0.24 ± 0.04 | 1.68 ± 0.09 | 0.37 ± 0.06 | 0.86 ± 0.13 | 0.19 ± 0.01 |
| Q2725 | 0.99 ± 0.08 | 0.15 ± 0.11 | 2.65 ± 0.12 | 0.41 ± 0.14 | 0.83 ± 0.14 | 0.13 ± 0.06 |
| Q2726 | 0.98 ± 0.15 | 0.16 ± 0.01 | 2.54 ± 0.03 | 0.41 ± 0.12 | 0.91 ± 0.11 | 0.15 ± 0.10 |
| Q2727 | 1.02 ± 0.07 | 0.17 ± 0.05 | 2.61 ± 0.15 | 0.44 ± 0.08 | 0.84 ± 0.02 | 0.14 ± 0.07 |
| Q2729 | 1.11 ± 0.05 | 0.18 ± 0.02 | 2.72 ± 0.11 | 0.43 ± 0.05 | 1.07 ± 0.07 | 0.17 ± 0.01 |
Data were averages of triplicate experiments, and errors represent standard deviation
A few approaches that engineered glyoxylate bypass for the production of glycolate from glucose in different microorganisms have been reported [13–15]. For examples, deletion of aceB and heterologous expression of ghrA from E. coli resulted in a titer of 4 g/L glycolate in Corynebacterium glutamicum with glucose as the sole carbon source. When the translation start codon ATG of isocitrate dehydrogenase (icd) was replaced by GTG, the production of glycolate was further increased to 5.3 ± 0.1 g/L [15]. A C. glutamicum strain was capable to produce glycolate by engineered glyoxylate bypass with glucose as the carbon source. The engineered E. coli strain EYX-2 had the highest reported glycolate titer of 56.44 g/L with 0.52 g/g glucose yield under 120 h fed-batch cultivation [14]. However, in our study, the titer of glycolate was only 0.51 ± 0.11 g/L via expression of aceA and ghrA and deletion of aceB. This titer was significantly lower than the previous results with glucose as carbon via the similar engineering. As shown in Fig. 1, the intermediate isocitrate can be both converted to α-ketoglutarate by isocitrate dehydrogenase (icd) and to glyoxylate by isocitrate lyase (aceA). The above two previous studies were all included the regulation of α-ketoglutarate branch. Therefore, our constructed route may redirect the carbon fluxes to glyoxylate synthesis insufficiently due to the α-ketoglutarate branch. Besides, the different carbon supply in our study may yield the different results as compared to the previous studies with glucose as the carbon even if the similar metabolic engineering is employed. The exact mechanism will be unravelled based on the metabonomic researches in future.
Production of glycolate from glycolaldehyde oxidation pathway
In this work, a new direct conversion from xylose to glycolate in an engineered E. coli was explored by introducing the xylose degradation pathway of C. crescentus. This new route for the bio-production of glycolate mainly consists of three parts (Fig. 1). Firstly, d-xylose was metabolized to d-xylonate by the catalysis of two heterogeneous enzymes Xdh and XylC from C. crescentus. Subsequently, d-xylonate was converted to glycolaldehyde by the E. coli native enzymes of xylonate dehydratase (yjhG) and 2-keto-3-deoxy-d-xylonate aldolase (yjhH) which has been identified in the earlier study [25, 26]. Finally, glycolaldehyde was oxidized to glycolate by the catalysis of aldehyde dehydrogenase (aldA). Overall, our constructed pathway was significantly different from the previous reported pathways for glycolate production from xylose [27, 28]. Stephanopoulos et al. demonstrated the assimilation of xylose via isomerization to xylulose, epimerization of xylulose to ribulose, and phosphorylation of the latter to yield ribulose-1-P, which was followed by an aldolytic cleavage to obtain glycolaldehyde and DHAP [27]. This pathway was similar to the work of Walther et al. In the constructed pathway of Walther et al. xylose was also assimilated to xylulose via isomerization. Then, xylulose was directly phosphorylated to xylulose-1-P and cleaved to glycolaldehyde and DHAP [28]. Though these two pathways had the same stoichiometry, the former additionally employed the synthetic xylulose epimerase reaction, which may complicate implementation of the pathway because another enzymatic activity has to be identified and optimized by protein engineering. However, in our designed pathway, d-xylose was oxidized to d-xylonate via two heterogeneous enzymes Xdh and XylC from C. crescentus. d-xylonate was then dehydrated to 2-dehydro-3-deoxy-d-xylonate, followed by an aldolytic cleavage to yield glycolaldehyde and pyruvate. The xylose assimilation in our designed pathway could produce one NADH via xylose oxidation step and did not depend on a phosphorylation step, which consumed one additional ATP per molecule of xylose. So, our pathway may have lower energy consumption. These advantages will be potential for the large-scale production of glycolate.
The engineered E. coli strain Q2562 was constructed by transformation of the two recombinant plasmids pLMe2 and pLMa2. The gel electrophoresis analysis was carried out for detecting the expressions of all the introduced genes. Figure 2 showed patterns of protein samples visualized by Coomassie Brilliant Blue staining. We noted distinct bands of the expected sizes from crude extracts of the recombinant strains when compared with the control strain. All the distinct bands of the proteins YjhH, XylC, Xdh, YjhG, AldA were found in Fig. 2 and confirmed that the introduced genes in different recombinant plasmids were expressed obviously.
Fig. 2.
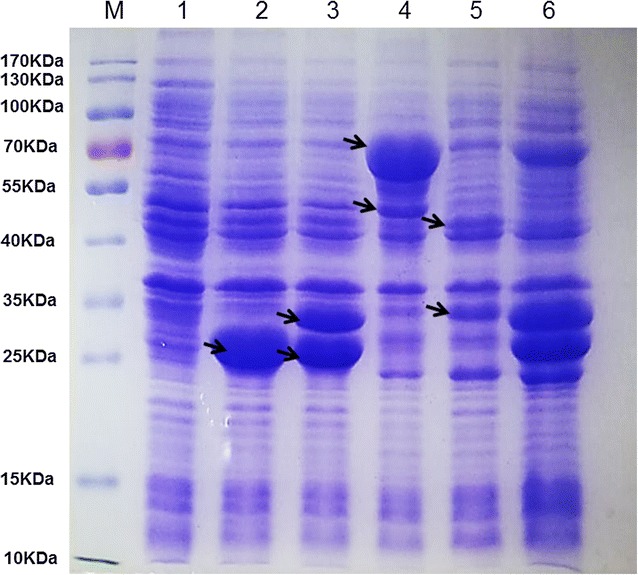
Coomassie brilliant blue-stained SDS-PAGE analysis of recombinant proteins from different constructs. Lane M, protein molecular weight marker; lane 1, crude cells extracts from E. coli BL21(DE3)/pETDuet1/pACYCDuet1; lane 2, crude cells extracts from E. coli BL21(DE3) strain harboring pETDuet1-yjhH; lane 3, crude cells extracts from E. coli BL21(DE3) strain harboring pETDuet1-xylC-xdh; lane 4, crude cells extracts from E. coli BL21(DE3) strain harboring pACYCDuet1-aldA-yjhG; lane 5, crude cells extracts from E. coli BL21(DE3) strain harboring pETDuet1-ghrA and pACYCDuet1-aceA; lane 6, crude cells extracts from E. coli BL21(DE3) strain harboring pETDuet1-yjhH-xylC-ghrA-xdh and pACYCDuet1-aldA-aceA-yjhG. The position corresponding to the overexpressed genes were indicated by an arrow
After cultivation of 24 h under shake-flask experiments, the glycolate production of Q2562 was 2.53 ± 0.08 g/L at the yield of 0.44 ± 0.10 g/g xylose, showing about ten times higher than those of Q2728 from the glyoxylate reduction pathway.
The cell biomass of the culture was 1.03 ± 0.16 gCDW/L, similar to the cell biomass as compared to that of Q2728 and Q2705 (Table 1). Considering all these results, a new route for the bio-production of glycolate from xylose was successfully established in E. coli. D-xylose was directly metabolized to D-xylonate by heterogenous enzymes Xdh and XylC from C. crescentus and then converted to glycolate by the native enzymes from E. coli. This research opened up a new prospect for bio-refinery of xylose and a new technology for the large-scale production of glycolate.
Production of glycolate from the two-combined pathway
In this new established glycolaldehyde oxidation pathway for the production of glycolate, the intermediate 2-keto-3-deoxy-d-xylonate was split to glycolaldehyde and pyruvate catalyzed by YjhH (Fig. 1). The yield of glycolate may be decreased due to the distribution of carbon flux to pyruvate. However, pyruvate could be converted to glycolate by a series of enzyme-catalyzed reactions from glyoxylate reduction pathway. Therefore, the glycolate yields may be improved by combining the glycolaldehyde oxidation pathway and glyoxylate reduction pathway. Genetic modifications of aceB deletion, aceA over-expression and ghrA over-expression were independently incorporated into strain Q2562, yielding new strains of Q2590, Q2725 and Q2726. The three strains were also cultivated in shake-flask with d-xylose as the sole carbon source. Surprisingly, Q2590 showed lower glycolate production and yield than those of Q2562. This result seems likely that the reduction of malate formation by aceB deletion reduces the formation of oxaloacetate which is necessary for the formation of isocitrate in the first place (Fig. 1). While Q2725 and Q2726 showed the similar glycolate productions and yields to those of Q2562 (Table 1). Due to the negative effects of aceB knockout on glycolate production, over-expressions of aceA and ghrA were chosen to incorporate into the strain Q2562 simultaneously, yielding a new strain Q2729. The engineered strain Q2729 reached 2.72 ± 0.11 g/L glycolate, higher than the value 2.53 ± 0.08 g/L of Q2562 (Table 1). However, the glycolate yield of Q2729 was similar to that of Q2562. Consistent with the above results, the glycolate production from xylose in glyoxylate reduction pathway was initially ineffective. Therefore, the glycolate productions and yields from the two-combined pathway were not significantly improved than those from the glycolaldehyde oxidation pathway.
The mutual transformation of glyoxylate and glycolate occurres in E. coli strains. Glyoxylate was reduced to glycolate by glyoxylate reductase (ghrA), and in turn, glycolate was oxidated to glyoxylate by glycolate oxidase (glcD) (Fig. 1). In order to avoid the conversion from glycolate to glyoxylate, the glcD gene was knocked out from Q2562, yielding strain Q2727. As shown in Table 1, the engineered strain Q2727 reached 2.61 ± 0.15 g/L glycolate, slightly higher than the value of 2.53 ± 0.08 g/L glycolate of Q2562. These results indicated that the reaction from glycolate to glyoxylate by GlcD had little effects on glycolate production.
Production of glycolate under fed-batch cultivation
In order to test the suitability of engineered strains for the larger-scale production of glycolate, the fed-batch cultivation was performed based on the results from the shake-flask cultures. The glycolate productions and yields of the engineered strains from two-combined pathway showed no obvious improvement than those of the strain Q2562 from glycolaldehyde oxidation pathway. So, the engineered strain Q2562 was performed for the fed-batch cultivation in a 3 L-scale laboratory fermenter. The OD600 and the concentrations of residual xylose, glycolate and acetate were monitored during the course of cultivation. The residual xylose concentration was controlled below 5 g/L. Figure 3 showed the time profiles of cell biomass, glycolate concentration and acetate concentration during the whole fermentation period, and all the three values increased gradually with time. After fermentation, the glycolate production of Q2562 was 28.82 ± 0.56 g/L and the glycolate yield was 0.38 ± 0.07 g/g xylose (Table 2). Importantly, the productivity of Q2562 was 0.60 ± 0.01 g/L/h, significantly higher than that of the value 0.47 g/L/h of the engineered E. coli strain EYX-2 which had the reported highest glycolate production from glucose. It was speculated that the strain Q2562 from glycolaldehyde oxidation pathway has the potential for large scale production of glycolate.
Fig. 3.
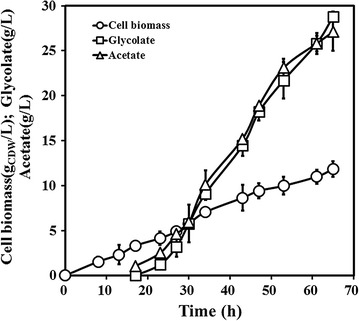
Time profiles for the cell biomass, glycolate production and acetate production of the engineered strain E. coli BL21(DE3)/pLMe2/pLMa2(Q2562) under fed-batch cultivation. Culture was performed in a 3 L laboratory fermenter containing 1 L of fermentation medium. Error bars represent ± standard deviation from the mean
Table 2.
Production of glycolate after 48 h of IPTG induction under fed-batch cultivation on minimal medium with d-xylose
| Strains | Cell biomass (gCDW/L) | Cell yield (gCDW/g xylose) | Glycolate concentration (g/L) | Glycolate yield (g/g xylose) | Acetate concentration (g/L) | Acetate yield (g/g xylose) | Glycolate productivity (g/L/h) |
|---|---|---|---|---|---|---|---|
| Q2562 | 11.84 ± 0.85 | 0.16 ± 0.02 | 28.82 ± 0.56 | 0.38 ± 0.07 | 27.18 ± 2.13 | 0.36 ± 0.03 | 0.60 ± 0.01 |
| Q2741 | 9.54 ± 0.76 | 0.11 ± 0.06 | 38.21 ± 0.86 | 0.42 ± 0.12 | 21.86 ± 1.31 | 0.24 ± 0.05 | 0.79 ± 0.01 |
| Q2742 | 9.25 ± 0.32 | 0.10 ± 0.01 | 43.60 ± 1.22 | 0.46 ± 0.03 | 9.08 ± 0.87 | 0.10 ± 0.10 | 0.91 ± 0.02 |
Data were averages of triplicate experiments, and errors represent standard deviation
To our surprise, Q2562 accumulated 27.18 ± 2.13 g/L acetate after cultivation. Acetate overflow was also occurred in fed-batch fermentation of engineered strain EYX-2, which was also about 20 g/L [14]. So, acetate overflow is one of the major problems for the large scale production of glycolate both from glucose to xylose. Acetate was produced from pyruvate by pyruvate oxidase (poxB) and produced from acetyl-CoA by phosphotransacetylase-acetate kinase (pta-ackA), respectively (Fig. 1). Approaches for overcoming acetate overflow may be beneficial for improving the glycolate production and xylose efficiency.
Overcoming acetate formation through iclR and ackA deletion
Acetate secretion has adverse effects on the cell growth, the macromolecules synthesis and the desirable metabolites formation [29, 30]. In order to overcome the acetate overflow, deletions of iclR and ackA were carried out. IclR (isocitrate lyase regulator) is a transcriptional regulator that represses gene expression of aceBAK operon. The aceBAK operon is responsible for coding the glyoxylate bypass enzymes of malate synthase (aceB), isocitrate lyase (aceA) and isocitrate dehydrogenase kinase/phosphatase (aceK) [31–33]. Glyoxylate bypass is the main pathway for acetate assimilation, and decreasing the gene expression of iclR may activate glycolate cycle and decrease acetate accumulation. AckA (acetate kinase) belongs to the acetate synthetic pathway. Deletion of ackA is a direct method to affect acetate formation [34]. The iclR and ackA were knocked out, yielding iclR mutant Q2280 and ackA mutant Q1951. The recombinant plasmids pLMe2 and pLMa2 were simultaneously transformed to Q2280 and Q1951, creating the engineered strain Q2741 and Q2742, respectively. Both the Q2741 and Q2742 strains were cultivated under fed-batch cultivation in a 3 L-scale laboratory fermenter. The results showed that knockout of iclR and ackA significantly improved the glycolate productions, productivities and yields (Table 2 and Fig. 4). The engineered strain Q2741 produced 38.21 ± 0.86 g/L glycolate, with 0.79 ± 0.01 g/L/h productivity and 0.42 ± 0.12 g/g xylose yield. Knockout of iclR improved the glycolate production by 1.32-fold than that of Q2562. Moreover, knockout of ackA showed better results for the above aspects. The engineered strain Q2742 produced 43.60 ± 1.22 g/L glycolate, showing 1.51-fold higher than that of Q2562. The productivity and yield reached 0.91 ± 0.02 g/L/h and 0.46 ± 0.03 g/g, respectively. This is the highest reported production of glycolate from xylose to the highest reported productivity to date.
Fig. 4.
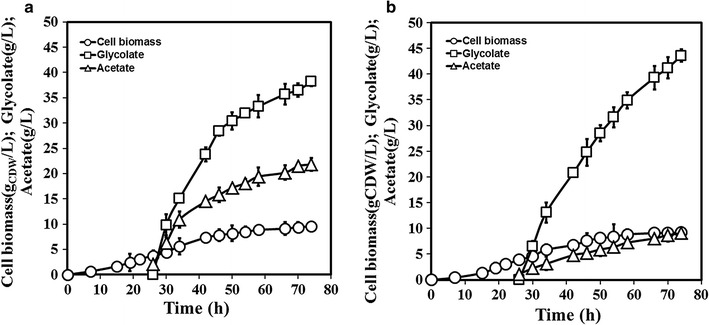
Knockout of iclR and ackA for overcoming acetate excretion. a Time profiles for the cell biomass, glycolate production and acetate production of iclR konckout strain E. coli BL21(DE3) iclR::Kan/pLMe2/pLMa2 (Q2741) under fed-batch cultivation. b Time profiles for the cell biomass, glycolate production and acetate production of the ackA knockout strain E. coli BL21(DE3) ackA::Kan/pLMe2/pLMa2 (Q2742) under fed-batch cultivation. Strains were cultivated in a 3 L laboratory fermenter containing 1 L of fermentation medium. Error bars represent ± standard deviation from the mean
In our designed glycolaldehyde oxidation pathway, xylose was metabolized to 2-keto-3-deoxy-d-xylonate and then cleaved to glycolaldehyde and pyruvate. Therefore, 1 mol glycolate could be produced from 1 mol xylose via the oxidation of intermediate glycolaldehyde. As this calculation, the maximum theoretical yield was about 0.51 g/g xylose. Obviously, the yield of engineering strain Q2742 approximated to the maximum theoretical yield through overcoming acetate formation. Compared to the work of Stephanopoulos et al. an engineered strain GA-10 produced 40 g/L glycolate, which presented the production comparable to that obtained by our study [27]. However, the productivity of our engineered strain Q2742 showed two times higher than that of the engineered strain GA-10. The increased productivity of the novel pathway may be of interest for the biotechnological production of glycolate.
Knockout of ackA showed dramatic effect on overcoming acetate overflow. The acetate concentration of Q2742 was decreased by 66%, from 27.18 ± 2.13 to 9.08 ± 0.87 g/L. It was only decreased by about 20%, from 27.18 ± 2.13 to 21.86 ± 1.31 g/L through iclR deletion. IclR is a transcriptional regulator that indirectly regulates acetate metabolism through aceBAK operon in glycolate bypass. By contrast, AckA is directly related to the acetate formation. It may be the reason for the phenomenon that ackA deletion showed more obvious effect on overcoming acetate overflow than iclR deletion. In this new constructed route for the production of glycolate, the intermediate 2-keto-3-deoxy-d-xylonate was split into pyruvate and glycolaldehyde by the catalysis of YjhH. Then the byproduct acetate was produced from pyruvate to its downstream acetyl-CoA (Fig. 1). Overcoming acetate formation through transcription regulations may improve the distribution of carbon flux for glycolate biosynthesis pathway and finally improve the glycolate production. In addition, the final cell biomass of Q2742 was much lower as compared to that of Q2562. The similar cases were also reported in some previous studies, suggesting that modifications of acetate metabolic pathway usually have negative impacts on cell growth [35–37]. A possible explanation is that ATP is released in the process of acetate production by Pta-AckA pathway. Disruption of the Pta-AckA pathway may affect ATP provision for the cell growth. In conclusion, the lower acetate yield and cell yield of Q2742 reflected the fact that cells regulate more carbon flux for glycolate production and it may be proved by the metabolomics research in future.
Conclusions
In this study, a new route (glycolaldehyde oxidation pathway) for biosynthesis of glycolate was established in E. coli by introducing the xylose degradation pathway of C. crescentus. d-xylose was directly metabolized to d-xylonate by heterogenous enzymes Xdh and XylC from C. crescentus and then converted to glycolate by the native genes from E. coli. In addition, ackA deletion successfully overcame the serious problem of acetate excretion during the process of glycolate production. The engineered strain Q2742 produced 43.60 ± 1.22 g/L glycolate, with 0.91 ± 0.02 g/L/h productivity and 0.46 ± 0.03 g/g xylose yield under fed-batch cultivation. The acetate concentration of Q2742 was decreased 66% by knocking out of ackA gene. In summary, this research opened up a new prospect for bio-refinery of xylose and established a new route for the biosynthesis of glycolate. The engineered strain Q2742 has the capacity for large scale production of glycolate and it could be widely used in industrial applications.
Methods
Plasmids and strains construction
Primers used for this study were listed in Table 3, plasmids and strains used in this study were listed in Table 4. E. coli DH5α was used for gene cloning and E. coli BL21(DE3) was used as the parent for all strain engineering. The xylC (GenBank Accession No.: NACL94328) and xdh (GenBank Accession No.: NACL94329) genes from C. crescentus were cloned into pETDuet1 vector between EcoRI, NotI and NdeI and KpnI sites to generate plasmid pETDuet1-xylC-xdh. Then yjhH (Gene ID: 948825) was cloned into pETDuet1-xylC-xdh vector between NcoI and EcoRI sites to create pETDuet1-yjhH-xylC-xdh (pLMe2). The recombinant plasmid pETDuet1-yjhH was also constructed under the same promoter to avoid the disturbance of Xdh and XylC with the similar molecular weight in protein expression and gel electrophoresis analysis. Finally, the ghrA (Gene ID 946431) was, respectively cloned into pETDuet1 and pLMe2 vectors between the same sites of NotI and AflII to create pETDuet1-ghrA (LMe1) and pETDuet1-yjhH-xylC-ghrA-xdh (pLMe3). The yjhG (Gene ID 946829) and aldA (Gene ID 945672) were cloned into pACYCDuet1 to generate pACYCDuet1-aldA-yjhG (pLMa2) vector between NdeI, XhoI and NcoI, EcoRI sites. The aceA (Gene ID 948517) was cloned into pACYCDuet1 and pLMa2 vectors using the same sites of EcoRI and HindIII to create pACYCDuet1-aceA (pLMa1) and pACYCDuet1-aldA-aceA-yjhG (pLMa3), respectively. The native E. coli genes of yjhH, aceA, ghrA and yjhG were generated by PCR using primers listed in Table 3. The chromosomal aceB, glcD, iclR and ackA genes of E. coli BL21 (DE3) strain encoded for malate synthase, glycolate oxidase, isocitrate lyase regulator and acetate kinase were knocked out via P1 vir-mediated transduction as previously described [38]. The donor strain JW3974, JW2946, JW3978 and JW2293 were purchased from the Keio collection [39]. The different plasmids were transformed to the corresponding hosts for the productions of glycolate by the glycolaldehyde oxidation pathway and/or glyoxylate reduction pathway.
Table 3.
Primers used in this study
| Name | Description |
|---|---|
| yjhH_F_NcoI | GGAATTCCATATGAAAAAATTCAGCGGCATTATTC |
| yjhH_R_EcoRI | CCGCTCGAGTCAGACTGGTAAAATGCCCT |
| xylC_F_ EcoRI | CCGGAATTCTAATACGACTCACTATAGGGGAATTG |
| xylC_R_NotI | AAGGAAAAAAGCGGCCGCTTAAACCAGACGAACTTCGTGCTG |
| ghrA_F_NotI | ATTTGCGGCCGCTCGCACAACGCTTTTCGGGA |
| ghrA_R_AflII | CCCACATGTTTAGTAGCCGCGTGCGCGGT |
| xdh_F_NdeI | GGGAATTCCATATG TCCTCAGCCATCTATCCC |
| xdh_R_KpnI | CGGGGTACC TCAACGCCAGCCGGCGTCGAT |
| aldA_F_NcoI | CCGCCATGGGCATGTCAGTACCCGTTCAACA |
| aldA_R_EcoRI | CCGGAATTCTTAAGACTGTAAATAAACCA |
| aceA_F_EcoRI | CCGGAATTCCACCACATAACTATGGAGCA |
| aceA_R_HindIII | CCCAAGCTTTTAGAACTGCGATTCTTCAG |
| yjhG_F_NdeI | GGAATTCCATATG TCTGTTCGCAATATTTTTGC |
| yjhG_R_XhoI | CCGCTCGAGTCAGTTTTTATTCATAAAATCGCG |
| ID-aceB_F | GTTATCAACAAGTTATCAAGT |
| ID-glcD_F | TCGATACTCTCTGCAACCAC |
| ID-iclR_F | CATAAAACGGATCGCATAACGC |
| ID-ackA_F | CATAAAACGGATCGCATAACGC |
| Kan_R | GGTGAGATGACAGGAGATCC |
Table 4.
Plasmids and strains used in this study
| Plasmids and strains | Description | Source |
|---|---|---|
| Plasmids | ||
| pETDuet1 | Amp r oriP BR322 lacI q P T7 | Novagen |
| pACYCDuet1 | Cm r oriP 15A lacI q P T7 | Novagen |
| pLMe1 | pETDuet1-ghrA | This study |
| pLMe2 | pETDuet1-yjhH-xylC-xdh | This study |
| pLMe3 | pETDuet1-yjhH-xylC-ghrA-xdh | This study |
| pLMa1 | pACYCDuet1-aceA | This study |
| pLMa2 | pACYCDuet1-aldA-yjhG | This study |
| pLMa3 | pACYCDuet1-aldA-aceA-yjhG | This study |
| Strains | ||
| E. coli DH5α | F− supE44 ΔlacU169 (ϕ80 lacZ ΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Invitrogen |
| E. coli BL21(DE3) | F− ompT gal dcm lon hsdSB (rB− mB−) λ(DE3) | Invitrogen |
| JW3974 | E. coli BW25113 aceB::kan | [39] |
| JW2946 | E. coli BW25113 glcD::kan | [39] |
| JW3978 | E. coli BW25113 iclR::kan | [39] |
| JW2293 | E. coli BW25113 ackA::kan | [39] |
| Q2589 | E. coli BL21(DE3) aceB::kan | This study |
| Q2723 | E. coli BL21(DE3) glcD::kan | This study |
| Q2280 | E. coli BL21(DE3) iclR::kan | This study |
| Q1951 | E. coli BL21(DE3) ackA::kan | This study |
| Q2728 | E. coli BL21(DE3)/pLMe1/pLMa1 | This study |
| Q2705 | E. coli BL21(DE3) aceB::kan/pLMe1/pLMa1 | This study |
| Q2562 | E. coli BL21(DE3)/pLMe2/pLMa2 | This study |
| Q2590 | E. coli BL21(DE3) aceB::kan/pLMe2/pLMa2 | This study |
| Q2725 | E. coli BL21(DE3)/pLMe2/pLMa3 | This study |
| Q2726 | E. coli BL21(DE3)/pLMe3/pLMa2 | This study |
| Q2727 | E. coli BL21(DE3) glcD::kan/pLMe2/pLMa2 | This study |
| Q2729 | E. coli BL21(DE3)/pLMe3/pLMa3 | This study |
| Q2741 | E. coli BL21(DE3) iclR::kan/pLMe2/pLMa2 | This study |
| Q2742 | E. coli BL21(DE3) ackA::kan/pLMe2/pLMa2 | This study |
Protein expression and gel electrophoresis analysis
For the expression of different recombinant proteins, single colonies of E. coli BL21 (DE3) harboring different recombinant plasmids were used to inoculate Luria–Bertani (LB) medium containing appropriate antibiotics and grown at 37 °C overnight. The saturated culture was diluted 1:100 into fresh LB medium and incubated under the same conditions. When the optical density at 600 nm (OD600) of the culture reached about 0.6, all the recombinant proteins expression were induced by 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG) for T7 promoter and growth was continued for 4 h at 30 °C. Cells were collected from 5 mL of bacteria cultures by centrifugation and washed with sterile distilled water. The washed pellets were suspended in 500 μL phosphate buffer saline buffer (pH 7.5) and subject to ultrasonication. The cell lysates were centrifuged and the supernatant obtained was mixed with 5× sodium dodecyl sulfate (SDS) sample buffer, heated at 100 °C for 10 min and then analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) according to standard protocols.
Shake-flask cultivation
To evaluate the ability of glycolate production in different engineered strains, shake-flask experiments were carried out in triplicate series of 250 mL Erlenmeyer flasks containing 50 mL medium with appropriate antibiotics. The medium comprises 14 g/L K2HPO4·3H2O, 5.2 g/L KH2PO4, 1 g/L NaCl, 1 g/L NH4Cl, 0.25 g/L MgSO4·7H2O, 0.2 g/L yeast extract. For the production of glycolate with xylose as the sole carbon source, E. coli strains were grown overnight at 37 °C with shaking in LB broth, and then 1:50 diluted into 50 mL medium with 15 g/L xylose. When the OD600 of the culture reached about 0.6, IPTG was added to a final concentration of 0.1 mM and further incubated 24 h at 30 °C for glycolate production. Samples were taken to determine the OD600, the concentrations of glycolate, by-product acetate and the residual xylose.
Fed-batch fermentation
In order to obtain higher glycolate concentrations, fed-batch cultures were performed in a Biostat B plus MO3L fermentor (Sartorius, Germany) containing 1 L of growth medium (9.8 g/L K2HPO4·3H2O, 2.1 g/L citric acid·H2O, 0.3 g/L ferric ammonium citrate, 3.0 g/L (NH4)2SO4) that was sterilized at 121 °C for 25 min. Xylose (20 g/L), MgSO4·7H2O (0.2 g/L) and 1000× trace elements (3.7 g/L(NH4)6Mo7O24·4H2O, 2.9 g/L ZnSO4·7H2O, 24.7 g/L H3BO3, 2.5 g/LCuSO4·5H2O, 15.8 g/L MnCl2·4H2O) were filter-sterilized separately and added prior to initiate the fermentation. 50 mL of seed culture was prepared by incubating the culture in shake flasks containing liquid LB medium overnight at 37 °C. During the fermentation process, continuous sterile air was supplied at a flow rate of 2 L/min. The pH was controlled at 7.0 during the whole fermentation via automatic addition of 25% (w/w) ammonia water, and the temperature was controlled at 37 °C. The dissolved oxygen (DO) level was maintained at 20 ± 1% by automatically adjusting stirring rate from 300 to 700 rpm. Antifoam was added to prevent frothing if necessary.
When the 20 g/L initial xylose was deleted, 60% (w/w) xylose was begun to feed into the fermentor to maintain the residual xylose below 5 g/L. The supply rate of 60% (w/w) xylose was adjusted according to the concentrations of residual xylose determined every 2 h. When the culture was reached to an OD600 of about 10, the temperature was switched to 30 °C, IPTG was added to the culture at a concentration of 0.1 mM and further incubated 48 h for glycolate production. Samples of the fermentation broth were removed at appropriate intervals to determine the OD600, the concentrations of glycolate and by-product acetate.
Analytic methods
Culture samples were taken at several times throughout the experiments. The cell concentration was assayed by measuring optical density of the culture at 600 nm (1 OD unit = 0.36 gCDW/L). (Cary 50 UV–Vis, Varian). Metabolites in the culture supernatant were analyzed by an Agilent 1200 Infinity series HPLC system equipped with an Aminex HPX-87H (Bio-Rad, Hercules, CA) column (300 × 7.8 mm). All samples were filtered through 0.22 μm syringe filter. Ultrapure water with 5 mM H2SO4 was used as the eluent at a flow rate of 0.5 mL/min. The oven temperature was maintained at 55 °C. Concentrations of xylose, glycolate and the by-product acetate were calculated based on standard curves.
Authors’ contributions
MX and ML developed the idea for the study. ML and GZ participated in the review of literature, and preparation of the manuscript. YD and HL helped to analyze the metabolites and residual xylose in the culture supernatant by HPLC and revised the manuscript. LM did the lab work, including the plasmids and strains construction, strain cultivation, fed-batch fermentation and products detection. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Ethics approval and consent to participate
Not applicable. The manuscript does not report date from humans or animals.
Funding
This work was supported by the National Natural Science Foundation of China (31670089 and 31722001), Project funded by China Postdoctoral Science Foundation (2016M600564) and Postdoctoral Applied Research Project of Qingdao.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- C. crescentus
Caulobacter crescentus
- E. coli
Escherichia coli
- IPTG
isopropyl β-d-thiogalactoside
- HPLC
high performance liquid chromatography
- PPP
pentose phosphate pathway
- TCA cycle
tricarboxylic acid cycle
- DO
dissolved oxygen
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
Contributor Information
Min Liu, Email: liumin@qibebt.ac.cn.
Yamei Ding, Email: elvadym@hotmail.com.
Mo Xian, Phone: +86-532-80662767, Email: xianmo1@qibebt.ac.cn.
Guang Zhao, Phone: +86-532-80662767, Email: zhaoguang@qibebt.ac.cn.
References
- 1.Wei G, Yang X, Gan T, Zhou W, Lin J, Wei D. High cell density fermentation of Gluconobacter oxydans DSM 2003 for glycolic acid production. J Ind Microbiol. 2009;36(8):1029–1034. doi: 10.1007/s10295-009-0584-1. [DOI] [PubMed] [Google Scholar]
- 2.Wu S, Fogiel AJ, Petrillo KL, Jackson RE, Parker KN, Dicosimo R, Ben-Bassat A, O’Keefe DP, Payne MS. Protein engineering of nitrilase for chemoenzymatic production of glycolic acid. Biotechnol Bioeng. 2008;99(3):717–720. doi: 10.1002/bit.21643. [DOI] [PubMed] [Google Scholar]
- 3.Rendon M, Colon L, Johnson L. Successful treatment of moderate to severe melasma with triple combination cream and glycolic acid peels. J ACAD Dermatol. 2008;58(2):2502–2506. [PubMed] [Google Scholar]
- 4.Kataoka M, Sasaki M, Hidalgo ARGD, Nakano M, Shimizu S. Glycolic acid production using ethylene glycol-oxidizing microorganisms. Biosci Biotechnol Biochem. 2001;65(10):2265–2270. doi: 10.1271/bbb.65.2265. [DOI] [PubMed] [Google Scholar]
- 5.Robertson GL. Food packaging: principles and practice. Packaging and Converting Technology. Boca Raton: CRC Press; 2006. pp. 1–550. [Google Scholar]
- 6.Fredenberg S, Wahlgren M, Reslow M, Axelsson A. The mechanisms of drug release in poly(lactic-co-glycolic acid)-based drug delivery systems—a review. Int J Pharm. 2011;415(1–2):34–52. doi: 10.1016/j.ijpharm.2011.05.049. [DOI] [PubMed] [Google Scholar]
- 7.John LD. Process for manufacture of glycolic acid. US2152852. 1939.
- 8.He YC, Xu JH, Su JH, Zhou L. Bioproduction of glycolic acid from glycolonitrile with a new bacterial isolate of Alcaligenes sp. ECU0401. Appl Biochem Biotechnol. 2009;160(5):1428–1440. doi: 10.1007/s12010-009-8607-y. [DOI] [PubMed] [Google Scholar]
- 9.Nieves LM, Panyon LA, Wang X. Engineering sugar utilization and microbial tolerance toward lignocellulose conversion. Front Bioeng Biotechnol. 2015 doi: 10.3389/fbioe.2015.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menon V, Rao M. Trends in bioconversion of lignocellulose: biofuels, platform chemicals & biorefinery concept. Prog Energ Combust. 2012;38(4):522–550. doi: 10.1016/j.pecs.2012.02.002. [DOI] [Google Scholar]
- 11.Zaldivar J, Nielsen J, Olsson L. Fuel ethanol production from lignocellulose: a challenge for metabolic engineering and process integration. Appl Microbiol Biotech. 2001;56(1–2):17–34. doi: 10.1007/s002530100624. [DOI] [PubMed] [Google Scholar]
- 12.Dyk JSV, Pletschke BI. A review of lignocellulose bioconversion using enzymatic hydrolysis and synergistic cooperation between enzymes-factors affecting enzymes, conversion and synergy. Biotechnol Adv. 2012;30(6):1458–1480. doi: 10.1016/j.biotechadv.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Koivistoinen OM, Kuivanen J, Barth D, Turkia H, Pitkänen JP, Penttilä M, Richard P. Glycolic acid production in the engineered yeasts Saccharomyces cerevisiae and Kluyveromyces lactis. Microb Cell Fact. 2013;12(1):1–16. doi: 10.1186/1475-2859-12-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng Y, Mao Y, Zhang X. Metabolic engineering of E. coli for efficient production of glycolic acid from glucose. Biochem Eng J. 2015;103:256–262. doi: 10.1016/j.bej.2015.08.008. [DOI] [Google Scholar]
- 15.Zahoor A, Otten A, Wendisch VF. Metabolic engineering of Corynebacterium glutamicum for glycolate production. J Biotechnol. 2014;192(11):366–375. doi: 10.1016/j.jbiotec.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 16.Lachke A. Biofuel from d-xylose—the second most abundant sugar. Resonance. 2002;7(5):50–58. doi: 10.1007/BF02836736. [DOI] [Google Scholar]
- 17.Kuhad RC, Gupta R, Khasa YP, Singh A, Zhang YHP. Bioethanol production from pentose sugars: current status and future prospects. Renew Sust Energ Rev. 2011;15(9):4950–4962. doi: 10.1016/j.rser.2011.07.058. [DOI] [Google Scholar]
- 18.Karhumaa K, Garcia SR, Hahnhägerdal B, Gorwagrauslund MF. Comparison of the xylose reductase-xylitol dehydrogenase and the xylose isomerase pathways for xylose fermentation by recombinant Saccharomyces cerevisiae. Microb Cell Fact. 2007;6(1):1–10. doi: 10.1186/1475-2859-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sonderegger M, Sauer U. Evolutionary engineering of Saccharomyces cerevisiae for anaerobic growth on xylose. Appl Environ Microb. 2003;69(4):1990–1998. doi: 10.1128/AEM.69.4.1990-1998.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wenning F. Metabolic studies on d-xylose, a pentose sugar. Xylose. 1956;68(12):248–251. [PubMed] [Google Scholar]
- 21.Stephens C, Christen B, Fuchs T, Sundaram V, Watanabe K, Jenal U. Genetic analysis of a novel pathway for d-xylose metabolism in Caulobacter crescentus. J Bacteriol. 2007;189(5):2181–2185. doi: 10.1128/JB.01438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephens Craig, Christen Watanabe, Kelly Fuchs, Thomas Jenal. Regulation of d-xylose metabolism in Caulobacter crescentus by a LacI-type repressor. J Bacteriol. 2007;189(24):8828–8834. doi: 10.1128/JB.01342-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meisenzahl AC, Shapiro L, Jenal U. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J Bacteriol. 1997;179(3):592–600. doi: 10.1128/jb.179.3.592-600.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hottes AK, Meewan M, Yang D, Arana N, Romero P, Mcadams HH, Stephens C. Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J Bacteriol. 2004;186(5):1448–1461. doi: 10.1128/JB.186.5.1448-1461.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H, Ramos KR, Valdehuesa KN, Nisola GM, Lee WK, Chung WJ. Biosynthesis of ethylene glycol in Escherichia coli. Appl Microb Biotechnol. 2013;97(8):3409–3417. doi: 10.1007/s00253-012-4618-7. [DOI] [PubMed] [Google Scholar]
- 26.Valdehuesa KNG, Liu H, Ramos KRM, Si JP, Nisola GM, Lee WK, Chung WJ. Direct bioconversion of d-xylose to 1,2,4-butanetriol in an engineered Escherichia coli. Process Biochem. 2014;49(1):25–32. doi: 10.1016/j.procbio.2013.10.002. [DOI] [Google Scholar]
- 27.Pereira B, Li ZJ, De MM, Giaw LC, Zhang H, Hoeltgen C, Stephanopoulos G. Efficient utilization of pentoses for bioproduction of the renewable two-carbon compounds ethylene glycol and glycolate. Metab Eng. 2016;34(6):80–87. doi: 10.1016/j.ymben.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Cam Y, Alkim C, Trichez D, Trebosc V, Vax A, Bartolo F, Besse P, François JM, Walther T. Engineering of a synthetic metabolic pathway for the assimilation of (d)-xylose into value-added chemicals. Acs Synth Biol. 2016;5:607–618. doi: 10.1021/acssynbio.5b00103. [DOI] [PubMed] [Google Scholar]
- 29.Farmer WR, Liao JC. Reduction of aerobic acetate production by Escherichia coli. Appl Environ Microb. 1997;63(8):3205–3210. doi: 10.1128/aem.63.8.3205-3210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cherrington CA, Hinton M, Chopra I. Effect of short-chain organic acids on macromolecular synthesis in Escherichia coli. J Appl Bacteriol. 1990;68(1):69–74. doi: 10.1111/j.1365-2672.1990.tb02550.x. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto K, Ishihama A. Two different modes of transcription repression of the Escherichia coli acetate operon by IclR. Mole Microb. 2003;47(1):183–194. doi: 10.1046/j.1365-2958.2003.03287.x. [DOI] [PubMed] [Google Scholar]
- 32.Gui L, Sunnarborg A, Pan B, Laporte DC. Autoregulation of iclR, the gene encoding the repressor of the glyoxylate bypass operon. J Bacteriol. 1996;178(1):321–324. doi: 10.1128/jb.178.1.321-324.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molina-Henares AJ, Krell T, Guazzaroni ME, Segura A, Ramos JL. Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors. FEMS Microb Rev. 2006;30(2):157–186. doi: 10.1111/j.1574-6976.2005.00008.x. [DOI] [PubMed] [Google Scholar]
- 34.Liu M, Feng X, Ding Y, Zhao G, Liu H, Xian M. Metabolic engineering of Escherichia coli to improve recombinant protein production. Appl Microb Biotechnol. 2015;99(24):10367–10377. doi: 10.1007/s00253-015-6955-9. [DOI] [PubMed] [Google Scholar]
- 35.De Mey M, De Maeseneire S, Soetaert W, Vandamme E. Minimizing acetate formation in E. coli fermentations. J Ind Microb Biotechnol. 2007;34(11):689–700. doi: 10.1007/s10295-007-0244-2. [DOI] [PubMed] [Google Scholar]
- 36.Dittrich CR, Vadali RV, Bennett GN, San KY. Redistribution of metabolic fluxes in the central aerobic metabolic pathway of E. coli mutant strains with deletion of the ackA-pta and poxB pathways for the synthesis of isoamyl acetate. Biotechnol Progr. 2005;21(2):627–631. doi: 10.1021/bp049730r. [DOI] [PubMed] [Google Scholar]
- 37.Contiero J, Beatty C, Kumari S, Desanti CL, Strohl WR, Wolfe A. Effects of mutations in acetate metabolism on high-cell-density growth of Escherichia coli. J Ind Microb Biotechnol. 2000;24(6):421–430. doi: 10.1038/sj.jim.7000014. [DOI] [Google Scholar]
- 38.Moore SD. Assembling new Escherichia coli strains by transduction using phage P1. Methods Mol Biol. 2011;765:155–169. doi: 10.1007/978-1-61779-197-0_10. [DOI] [PubMed] [Google Scholar]
- 39.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2(1):8. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.


