Figure 3.
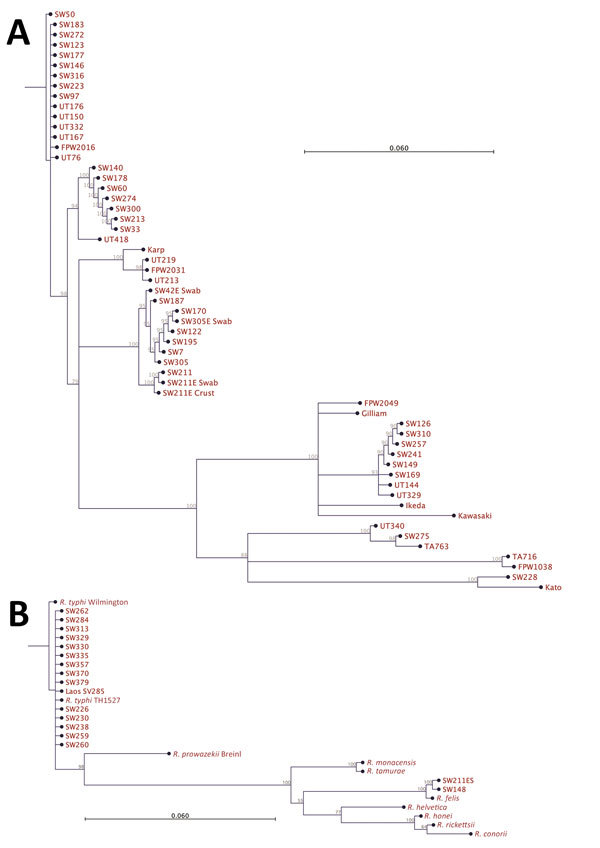
Phylogenetic analysis of pathogens contributing to rickettsial illnesses, Chittagong Medical College Hospital, Chittagong, Bangladesh, August 2014–September 2015. A) Phylogenetic dendrogram based on the nucleotide sequence of the partial open reading frame of the 56-kDa TSA gene (aligned and cropped to ≈450 bp), depicting Orientia tsutsugamushi strains in relationship with reference and other strains. O. tsutsugamushi genotypes in Bangladesh included Karp, Gilliam, Kato, and TA763 strains, with a predominance of Karp-like strains. B) Rickettsia spp. as characterized by 17-kDa gene sequencing (aligned and cropped to 314 bp). The predominant pathogen identified was R. typhi. Of note, 2 R. felis infection cases were identified, including 1 systemic bloodstream infection and 1 scrub typhus case with eschar swab positivity in patient no. SW211ES (Technical Appendix Table), whose blood specimen was negative for R. felis, suggesting possible skin colonization of R. felis. Scale bar indicates nucleotide substitutions per site; branches shorter than 0.002 are shown as having a length of 0.002.
