Figure.
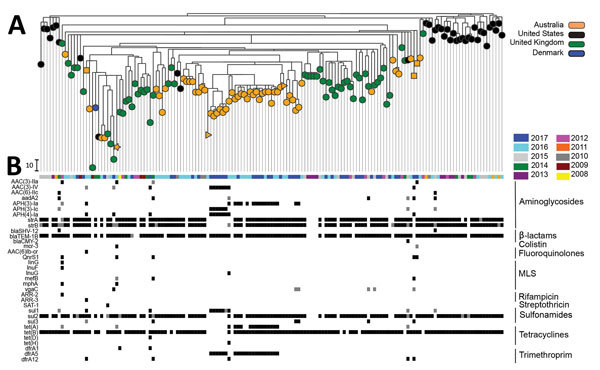
Maximum-likelihood phylogeny of whole-genome single-nucleotide polymorphisms (SNPs) of 153 Salmonella enterica 4,[5],12:i:- sequence type (ST) 34 isolates and acquired drug-resistance genes. A) SNP analysis was conducted by performing whole-genome alignment of ST34 isolates from New South Wales (NSW), Australia, and a selection of published ST34 isolates collected in the United Kingdom, United States, and Denmark by using Snippy Core (https://github.com/tseemann/snippy) (Technical Appendix). Regions of recombination were identified by using BratNextGen (www.helsinki.fi/bsg/software/BRAT-NextGen/) and removed. SNPs were identified by using SNP-sites (https://github.com/sanger-pathogens/snp-sites), and the phylogeny was generated by using FastTree (www.microbesonline.org/fasttree/). Phylogeny and antimicrobial resistance metadata were combined by using Microreact (https://microreact.org/showcase). The colistin-resistant ST34 isolate from NSW is denoted by an orange star, fluoroquinolone-resistant isolates from NSW by orange squares, and pork isolates from NSW by orange triangles. Scale bar indicates 10 SNPs. B) Year of isolation and acquisition of drug resistance. Acquired drug-resistance genes were identified by screening all isolate contigs through the ResFinder (8) and CARD (https://card.mcmaster.ca/) databases by using ABRicate version 0.5 (https://github.com/tseemann/abricate). Only genes with a 100% homology match in >1 isolate are shown. Columns depict the results for individual isolates; rows represent acquired drug-resistance genes. The antibiotic class that genes confer resistance against is indicated at right. White indicates that the specified gene was not detected, gray indicates that the specified gene was detected but sequence homology against the reference was <100%, black indicates a perfect match between the isolate and reference gene sequence. MLS, macrolide, lincosamide, and streptogramin B.
