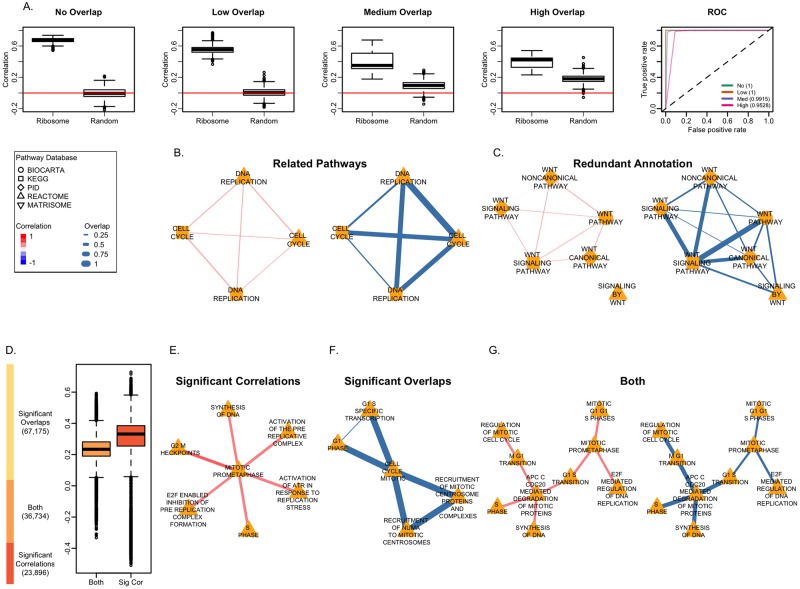
Fig 2. Significant correlations between the ribosome pathway and impact of gene overlap.
(A) Boxplots of the correlation estimates between the Ribosome gene sets and random gene sets, and receiver operating characteristic (ROC) curves with the corresponding area under the curve (AUC) values in parenthesis under different degrees of overlap: no overlap, low overlap (overlap coefficient 0.0469, AUC = 1), medium overlap (overlap coefficient 0.5517, AUC = 0.9915) and high overlap (overlap coefficient 0.8532, AUC = 0.9528). The shape of the node in the following networks corresponds to the pathway database. For coexpression networks, the edge color indicates the value of the correlation and edge width is proportional to the correlation magnitude. For the overlap networks, the edge width is proportional to the overlap coefficient. (B) Pathway coexpression and overlap network for the KEGG and Reactome annotations of the Cell Cycle and DNA Replication pathways. These pathways have related functions and share genes between them. (C) Pathway coexpression network and overlap network for different versions of the Wnt Signaling pathway. In the coexpression network, missing edges correspond to correlations that are not significant. These pathway annotations are redundant and represent the same function (D) The stacked bar plot shows the number of pathways pairs with only significant correlations in red, with only significant overlaps in yellow, and with both in orange. The boxplots show the distribution of the correlation coefficients with pathway pairs with only significant correlations (red) and with both significant overlaps and significant correlations (orange). (E) Pathway coexpression network for the Reactome pathways related to the mitotic metaphase of the cell cycle with significant correlations but no shared genes. (F) Overlap network for Reactome pathways related to the mitotic cell cycle with significant overlaps but no significant correlations. (G) Pathway coexpression network and overlap network for cell cycle phases and related processes from Reactome with both significant correlations and significant overlaps.