Abstract
Phagocytosis of dying cells is critical in development and immunity1–3. While proteins for recognition and engulfment of cellular debris following cell death are known4,5, proteins that directly mediate phagosome sealing are uncharacterized. Furthermore, whether all phagocytic targets are cleared using the same machinery is unclear. Degeneration of morphologically-complex cells, such as neurons, glia, and melanocytes, produces phagocytic targets of various shapes and sizes located in different microenvironments6,7. Such cells, therefore, offer unique settings to explore engulfment program mechanisms and specificity. Here we report that dismantling and clearance of a morphologically-complex C. elegans epithelial cell requires separate cell-soma, proximal-, and distal-process programs. Similar compartment-specific events govern elimination of a C. elegans neuron. While canonical engulfment proteins drive cell-soma clearance, these are not required for process removal. We find that EFF-1, a protein previously implicated in cell-cell fusion8, specifically promotes distal-process phagocytosis. EFF-1 localizes to phagocyte pseudopod tips, and acts exoplasmically to drive phagosome sealing. eff-1 mutations result in phagocytosis arrest with unsealed phagosomes. Our studies suggest universal mechanisms for dismantling morphologically-complex cells, and uncover a phagosome sealing component promoting cell-process clearance.
The C. elegans tail-spike cell is a morphologically-complex cell that extends a microtubule-laden process to animal’s tail tip. Wrapped around the tail-spike-cell process is the hyp10 epithelial cell, which also extends posteriorly (Fig. 1a–d; Supplementary Fig 1a). Ectopic tail-spike cell generation results in a forked tail (n=5; Supplementary Fig.1b,c;9); while early tail-spike-cell ablation perturbs tail morphogenesis (5/5 ablated animals; Supplementary Fig. 1d,e). Thus, like hyp10, the tail-spike cell plays a key role in C. elegans tail morphogenesis.
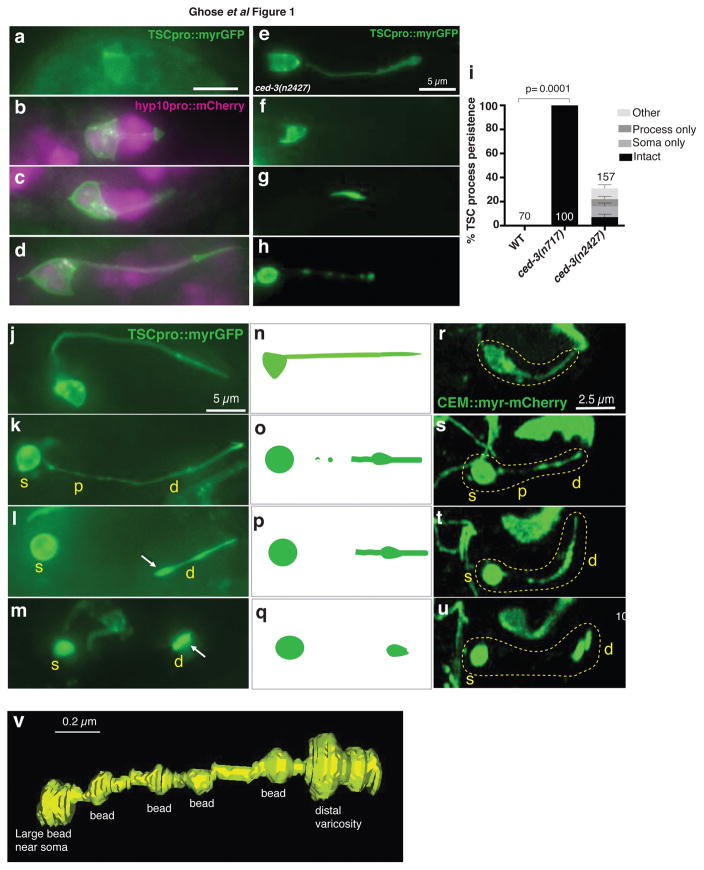
Figure 1. The tail-spike cell (TSC) and CEM neurons undergo a similar degeneration sequence.
(a–d) TSC in comma stage, 1.5 fold, 2-fold, and 3-fold embryos, respectively and its association with hyp10. n=5 biologically independent animals each with similar results. Scale bar: 5 μm. (e–h) ced-3(n2427) mutants exhibit an intact cell, persistent soma only, persistent process only, or intermediate degeneration (other), respectively. n=10 biologically independent animals each with similar results. Scale bars: 5 μm. (i) TSC persistence in ced-3 mutants. Data are mean +/− s.e.m. Statistics: two-tailed unpaired t-test. Individual p values: see Supplementary Table 2. Numbers inside/outside bars, total animals scored per genotype. Data are from 3 independent scoring experiments. n=sample sizes for statistics are as follows: WT:70, ced-3(n717):100, ced-3(n2427): 157 (other: 42, process only: 31, soma only:47, intact: 37. (j) TSC in 3-fold embryo. (k) TSC soma rounding and proximal process beading. (l,m) distal process retraction. n=10 biologically independent animals each. Scale bar: 5 μm. (r–u) Stills from movies showing CEM death. Scale bar: 2.5 μm. Time: hours:minutes post-fertilization, 07:40 (r), 08:05 (s), 08:15 (t), 08:30 (u). (n–q) schematics for j–m and r–u. n=6 biologically independent animals each. Yellow text: s, soma; p, proximal process; d, distal process. White arrowhead, distal varicosity. (v) 3D reconstruction of a degenerating TSC in an eff-1(ns634) animal. TSC death stages in this mutant are similar to WT (Main text, Supplementary Movie 5). Reconstruction based on Supplementary Movie 2. Scale bar: 0.2 μm. Statistics source data are provided in Supplementary Table 2.
Once tail formation is complete, the tail-spike cell dies through transcriptional induction of the main C. elegans caspase, CED-310. By following myristoylated-GFP expressed in the tail-spike cell, we found that a strong ced-3 loss-of-function mutation promotes tail-spike cell soma and process persistence in larvae (Supplementary Fig. 1d, Fig. 1i;10). Only 30% of animals carrying a weak ced-3 allele, however, exhibit tail-spike cell persistence. Of these animals, 24% exhibit a fully intact cell, 30% have an intact cell soma alone, and 18% exhibit an intact cell process alone. Remaining animals display tail-spike cells at various states of degeneration (Fig. 1a–i). These observations demonstrate that CED-3 caspase drives cell-process and cell-soma degeneration independently.
To examine this idea more closely, we used myristoylated-GFP to follow tail-spike-cell death dynamics in 3-fold stage embryos by taking still images of different embryos at different time points (n>50). We find that degeneration begins with beading of the proximal cell process and rounding of the cell soma, followed by appearance of a varicosity in the distal process. Clearance of the proximal process ensues, followed by distal process retraction into the distal varicosity (Fig. 1j–q). Tail-spike cell body and the distal varicosity are then engulfed and cleared by different neighboring cells, with hyp10 engulfing the varicosity (see below). To confirm this event sequence, we imaged 14 individual animals over time using a custom-built iSPIM light-sheet microscope, which acquires rapid image volumes without motion-induced blurring11. We found the same sequence of events occurring over a period of about 170 minutes (Supplementary Movie 1). Corroborating these results, serial-section TEM of a 3-fold embryo reveals proximal beading and distal varicosity formation (Supplementary Movie 2; Fig. 1v). We also imaged a myristoylated mCherry reporter and obtained similar results (Supplementary Fig 1f–i). Thus, independent morphological and molecular events dismantle the tail-spike cell soma, proximal-, and distal process.
To determine whether similar degeneration dynamics occur in other morphologically-complex cells, we examined the sex-specific CEMVL neuron that dies in hermaphrodites, but survive in males12. As embryonic CEMVL reporters are not known, we labeled the cells using our recently-developed gene-induction system, in which CEMVL precursor cells, in animals carrying a heat-shock-promoter::mCherry construct, are heated with an infrared laser for cell-specific labeling13. Time-lapse movies reveal that once generated, the CEMVL neuron extends a dendrite towards the nose tip. Dismantling of this axon-less neuron then occurs is a sequence resembling that of tail-spike cell dismantling (n=6; Fig. 1r–u; Supplementary Movie 3). Thus, two different morphologically-complex cell types employ common spatially-restricted programs for death and clearance. Importantly, these studies also shed light on dendrite destruction, a phenomenon much less understood than axon degeneration14.
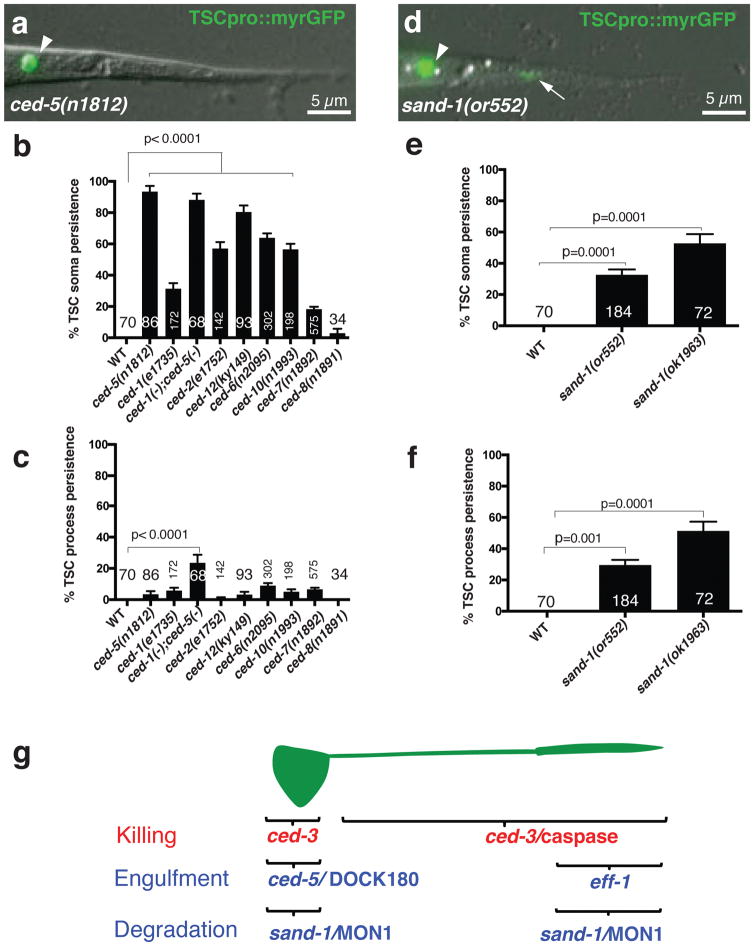
We next wondered whether the compartment-specific clearance events we uncovered correspond to distinct molecular programs. As imaging of CEMVL neurons is cumbersome, we pursued these studies in the tail-spike cell. We found that mutations affecting CED-1/Draper/MEGF10 and other apoptotic corpse engulfment regulators disrupt tail-spike cell soma clearance, with loss of CED-5/Dock180 having the strongest effect (Fig. 2a,b;15,16). However, single mutants in these genes, or a ced-1; ced-5 double mutant have only minor defects in proximal or distal process clearance (Fig. 2c). Mutations in sand-1/MON1, required for phagosome maturation17, block cell soma and distal process clearance, but have no effects on proximal process elimination (Fig. 2d–f; Supplementary Movie 4). These results suggest that distinct molecular programs drive tail-spike cell soma, proximal-, and distal process dismantling (Fig. 2g), and that additional genes promoting cell-soma and process clearance remain to be identified.
Figure 2. Canonical engulfment genes are not required for TSC process clearance.
(a) Unengulfed soma in ced-5(n1812) L1 animal. n=86 biologically independent animals with similar results. (b) TSC soma clearance defects. (c) TSC process clearance defects. n=sample sizes for statistics are as follows, with n referring to number of biologically independent animals.WT: n=70, ced-5(n1812): n=86, ced-1(e1735): n=172, ced-1(-);ced-5(-):n=68, ced-2(e1752):n=142, ced-6(n2095):n=302, ced-10(n1993):n=198, ced-7(n1892):n=575, ced-8(n1981): n=34. (d) Undegraded soma and process in sand-1(or552) L1 animal. n=184 biologically independent animals with similar results. (e,f). TSC soma, process degradation defects in sand-1 mutants, respectively. n=sample sizes for statistics are as follows, with n referring to number of biologically independent animals.WT:n=70, sand-1(or552):n=184), sand-1(ok1963):n=72 (g). Genes promoting TSC regional killing and clearance. Arrowhead, soma; arrow, process. Data are mean +/− s.e.m. Statistics: two-tailed unpaired t-test. Individual p values: see Supplementary Table 2. Numbers inside bars, total animals scored per genotype. 3 independent scoring experiments were done. Scale bar: 5 μm. Statistics source data are provided in Supplementary Table 2.
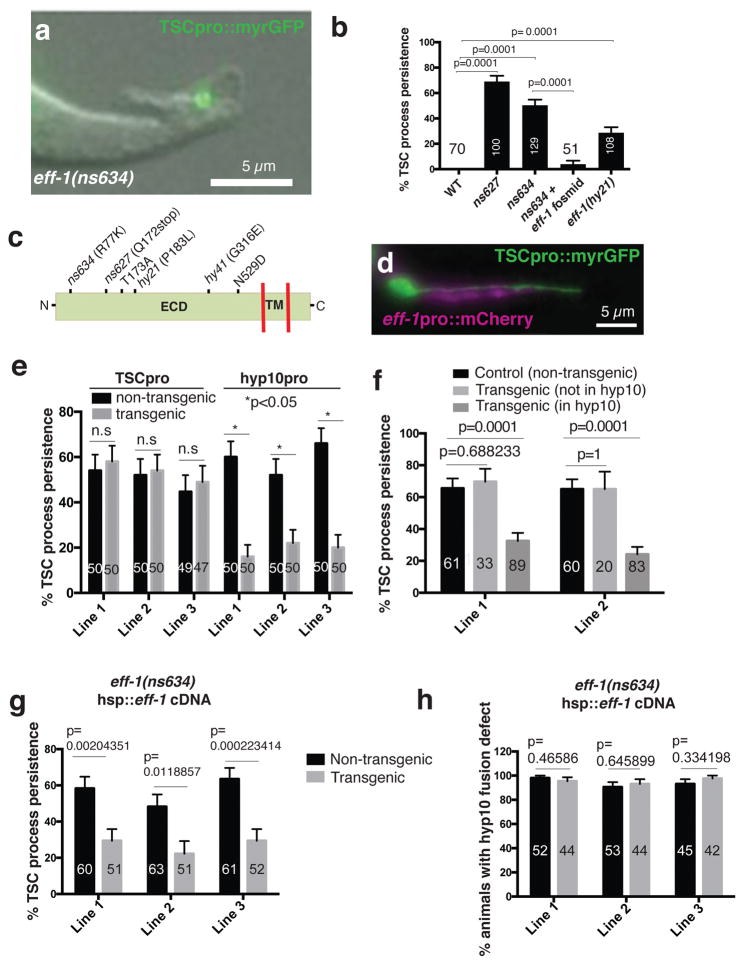
To seek such genes, we mutagenized C. elegans expressing myristoyl-GFP in the tail-spike cell, and screened through ~27,000 F2 progeny for mutants with persisting tail-spike cell fragments. In addition to mutations in ced-3/caspase and sand-1/MON1, we isolated two mutants, ns627 and ns634, that exhibit process clearance defects (Fig. 3a,b), but do not affect soma removal (n>100 each). Mutant L1 larvae do not exhibit other persistent cell corpses seen in apoptotic engulfment mutants18 (n>70).
Figure 3. EFF-1 fusogen mediates distal process clearance.
(a) L1 eff-1(ns634) mutant. n=129 biologically independent animals with similar results. (b) Process clearance defects in indicated genotypes. n=sample sizes for statistics are as follows with n referring to number of biologically independent animals.WT: n=70, ns627: n=100, ns634:n=129, ns634+ fosmid: n=51, eff-1(hy21):n=108. (c) EFF-1 structure and mutation sites. ECD, extracellular domain; TM, transmembrane domain. hy41 was previously identified14. (d) ced-3(n717) mutant showing hyp10 eff-1 expression. n=4 biologically independent animals with similar results. (e) eff-1 mutant cell-specific rescue. n=sample sizes for statistics are as follows with n referring to number of biologically independent animals: TSC Line 1: n=50 (non transgenic), n=50 (transgenic); TSC Line 2: n=50 (non transgenic), n=50 (transgenic); TSC Line 3: n=49 (non transgenic), n=47 (transgenic); hyp10 Lines 1–3 each: n=50 (non transgenic), n=50 (transgenic) (f) Mosaic analysis showing EFF-1 function in hyp10 for TSC distal process clearance. n=sample sizes for statistics are as follows, Line 1: Control: 61, Transgenic (not in hyp10): n=33, Transgenic (in hyp10): n=89; Line 2: Control: n=60, Transgenic (not in hyp10): n=20, Transgenic (in hyp10): n=83. (g) Rescue of eff-1 mutant TSC clearance defect with heat-shock promoter::eff-1 cDNA following heat exposure. n=sample sizes for statistics are as follows, Line 1: n=60 (non-transgenic), n=51 (transgenic); Line 2: n=63 (non-transgenic), n=51 (transgenic); Line 3: n=61 (non-transgenic), n=52 (transgenic). (h) Same as g, except rescue of fusion examined. n=sample sizes for statistics are as follows, Line 1: n=52 (non-transgenic), n=44 (transgenic); Line 2: n=53 (non-transgenic), n=44 (transgenic); Line 3: n=45 (non-transgenic), n=42 (transgenic). Data are mean +/− s.e.m. Statistics: two-tailed unpaired t-test. n.s., non significant (p>0.05). Individual p values: see Supplementary Table 2. Numbers inside bars, total animals scored per genotype. For rescue experiments, 2–3 independent transgenic lines were scored. For other experiments, 3 independent scoring experiments were done. For heat-shock rescue, scoring was done 2 hours after heat-shock. Scale bars: 5 μm. Statistics source data are provided in Supplementary Table 2.
Using whole-genome sequencing, single-nucleotide-polymorphism mapping, and transformation rescue, we demonstrated that both mutants harbor causal lesions in the gene eff-1, encoding a transmembrane protein with structural homology to viral class II fusion proteins, and required for cell-cell fusion8. Confirming this, animals homozygous for the canonical eff-1(hy21) allele also exhibit tail-spike cell clearance defects (Fig. 3b). Both mutants we isolated have animal morphology defects resembling those of eff-1(hy21) mutants, suggesting that they also lack cell fusion events. Consistent with this, both alleles perturb the EFF-1 extracellular domain (Fig. 3c).
To determine which part of the tail-spike cell process persists in eff-1 mutants, we imaged tail-spike cell death using iSPIM. While cell soma degradation, proximal process degradation, and distal process retraction are unaffected, distal process varicosity clearance is blocked (Supplemental Movie 5).
To determine where EFF-1 functions, we first examined its expression pattern. In ced-3(n717) mutant larvae, in which a live tail-spike cell persists, an eff-1 promoter::mCherry reporter is expressed in hyp10, but not in the tail-spike cell (Fig. 3d). Thus, EFF-1 likely functions in the engulfing cell. Supporting this, expression of an eff-1 cDNA using a tail-spike-cell-specific promoter fails to rescue distal-process clearance defects of eff-1 mutants; however, expression of the same cDNA using a hyp10-expressed eff-1 promoter fragment rescues these defects (Fig. 3e). Furthermore, in animals carrying an unstable extrachromosomal array containing the eff-1 locus, array presence/absence in hyp10 strongly correlates with distal fragment clearance/persistence (Fig. 3f). Thus, EFF-1 functions in the hyp10 engulfing cell for tail-spike-cell distal-process clearance (Fig. 2g).
During embryogenesis, two hyp10 cells fuse to form the hyp10 syncytium, which engulfs the tail-spike cell distal process. Since EFF-1 controls cell fusion, distal process clearance defects in eff-1 mutants may therefore reflect hyp10 cell-cell fusion failure. Arguing against this, however, eff-1 mutants have fully penetrant tail-morphology defects, likely resulting from cell-cell fusion defects; yet we found rare eff-1(ns634); hyp10p::eff-1 animals in which tail tip defects were rescued, but distal process engulfment still failed, suggesting that hyp10 cell-cell fusion and distal-fragment clearance are separable events. To test this directly, we followed hyp10 cell-cell fusion using the AJM-1::GFP reporter, labeling junctions between unfused cells19. 100% of eff-1 mutant animals exhibit hyp10 cell-cell fusion defects; however, only 40% have tail-spike-cell clearance defects (n=48) (Supplementary Fig. 2b,c). Thus, blocking hyp10 cell-cell fusion is not sufficient to block distal process engulfment. To further address this issue, we subjected eff-1(ns634) mutants, carrying heat-inducible eff-1 cDNA transgenes, to elevated temperatures, and assessed distal-process clearance and hyp10 cell-cell fusion. While distal fragments were cleared within two hours of heat exposure, cell fusion was restored more slowly (Fig. 3g,h), beginning only 4–6 hours later. Together, these studies suggest that hyp10 cell-cell fusion defects alone cannot explain tail-spike cell distal fragment clearance failure. Thus, eff-1 likely has direct roles in distal-process clearance.
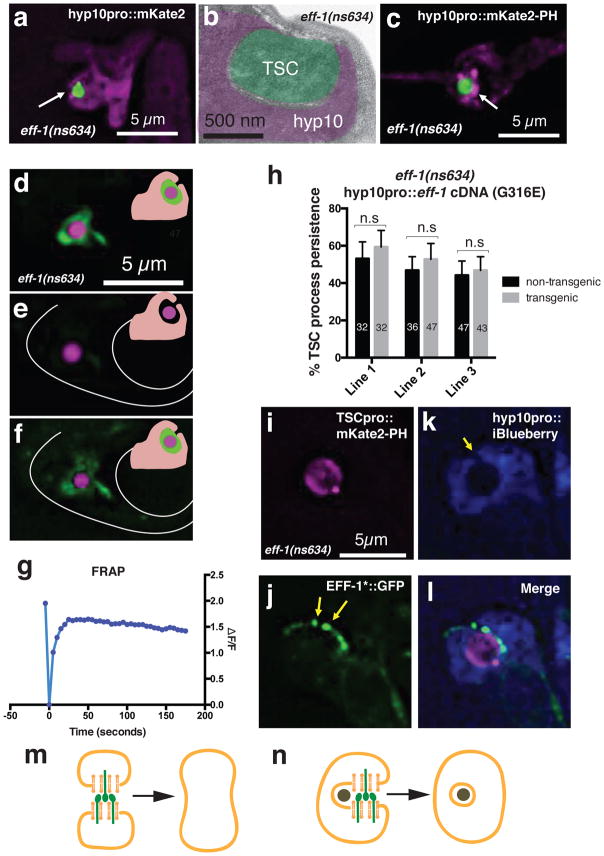
EFF-1 could be required for distal-process varicosity recognition by hyp10, phagosome sealing, phagosome maturation, or lysosomal degradation. We found that the varicosity is usually surrounded by hyp10 pseudopods in eff-1 mutants; however, gaps are often seen in optical cross sections (Fig. 4a; Supplementary Movie 6), suggesting that while the varicosity is being recognized, it is not fully internalized. Electron micrographs of an eff-1(ns634) mutant support the idea that phagosome sealing is defective in eff-1 mutants (Fig 4b).
Figure 4. EFF-1 promotes phagosome sealing.
(a) TSC distal process incompletely engulfed by hyp10 cell. Arrow, phagosome opening. n=15 biologically independent animals with similar results. (b). Electron micrograph of eff-1(ns634) L1 showing open phagosome. Area not pseudocolored, cuticle. n=1 animal. (c) Enrichment of open phagosome marker (arrow) around TSC fragment. n=10 biologically independent animals with similar results. (d–f) FRAP experiment in eff-1(ns634); cup-2(ar506); ssGFP: green, secreted GFP; magenta, TSC; inset, secreted GFP (green) surrounding TSC in phagosome space. (d) before, (e) immediately after, (f) 30 minutes after photobleaching. n=11 biologically independent animals with similar results. (g) Quantification of a different FRAP experiment. n=19 biologically independent animals with similar results. (h) EFF-1(G316E) cannot rescue eff-1(ns634) distal fragment clearance defect. n=sample sizes for statistics are as follows, Line 1: n=32 (non-transgenic), n=32 (transgenic); Line 2: n=36 (non-transgenic), n=47 (transgenic); Line 3: n=47 (non-transgenic), n=43 (transgenic). (i–l) Co-localization of EFF-1 (arrowheads) and phagosome opening (arrow). n=4 biologically independent animals with similar results. (m,n) Models of EFF-1 (green) function in cell-cell fusion and phagosome sealing, respectively. Data are mean +/− s.e.m. Statistics: two-tailed unpaired t-test. Individual p values: see Supplementary Table 2. n.s., not significant (p>0.05). Numbers inside bars, total animals scored per genotype. 3 independent transgenic lines were scored for rescue experiment. FRAP experiments were repeated 38 times. Scale bars: 5 μm.
To confirm these results, we examined localization of phagosome-associated proteins around the distal fragment remnant. We found that hyp10-expressed mKate2 fluorescent protein fused to the PH domain of PLCδ, which marks unsealed phagosomes20, is enriched around the tail-spike-cell distal-process varicosity (8/8 animals Fig. 4c; Supplementary Movie 7). The closed phagosome marker GFP::2xFYVE20 is generally not enriched (19/20 animals; Supplementary Fig. 3a–c; Supplemental Movie 8). A marker for mature phagosomes, RAB-721, also does not localize around the distal-process remnant (Supplementary Fig. 3d–f; Supplementary Movie 9); nor does LAAT-1::mCherry, a phagolysosome marker20 (Supplementary Fig. 3g–i; Supplementary Movie 10). Thus, the distal-process fragment resides in an unsealed phagosome.
To directly test whether the distal-process phagosome is open in eff-1 mutants, we reasoned that its interior should be continuous with the extracellular milieu. A fluorescent molecule expressed in this milieu should penetrate the phagosome only if the organelle is open (Supplementary Fig. 4e,f). We therefore generated eff-1(ns634); cup-2(ar506) mutants expressing secreted GFP (ssGFP) from body muscle using the myo-3 promoter22. In wild-type animals, ssGFP is taken up by coelomocyte scavenger cells; however, cup-2(ar506) blocks this uptake, allowing ssGFP extracellular accumulation. cup-2(ar506) mutants do not have tail-spike-cell clearance defects (n=10). Most eff-1(ns634); cup-2(ar506); myo-3p::ssGFP animals exhibit GFP fluorescence between the phagosome membrane and the tail-spike cell remnant, suggesting access to the phagosome at some time (Fig. 4d; Supplementary Fig 4a–d; Supplementary Movie 11). We photobleached the signal surrounding the tail-spike cell remnant and assessed whether fluorescence recovered (Supplementary Fig 4). Signal recovery occurred over a period of 5 seconds (19 animals) to 30 minutes (11 animals) in most animals observed (n=38; Fig. 4d–f; Supplementary Movie 12, 13), strongly supporting the notion that the mutant phagosome is unsealed. We speculate that fluorescence recovery time may correlate with phagosome gap size. As a control, we looked at sand-1(or522);cup-2(ar506) mutants (n=45), in which a sealed, persisting phagosome is expected. 44 animals had no GFP accumulation around the TSC, likely because phagosomes sealed prior to ssGFP expression. One animal exhibited GFP accumulation (Supplementary Fig. 4g–r; Supplementary Movie 14), but GFP fluorescence did not recover following photobleaching, suggesting the phagosome is closed (Supplementary Fig 4f).
A number of C. elegans proteins are implicated in phagosome sealing, including MTM-1/myotubularin, OCRL-1/Inositol 5-phosphatase, LST-4/SNX9, PIKI-1/PI3 kinase, and DYN-1/dynamin20. Dynamin proteins also play roles in intracellular scission of endocytic vesicles, although dynamin-independent endocytosis has been documented23. Mutants in these genes, however, have no effect on distal process removal (Supplementary Table 1). We also examined DYN-1::GFP localization using iSPIM in wild-type animals (n=3 movies). While DYN-1::GFP enrichment is seen around the cell soma, enrichment is not observed around the distal process fragment. Thus, phagosome sealing during distal process engulfment may be independent of known sealing components.
EFF-1 roles in cell fusion have been extensively characterized. Like viral fusion proteins, expression of EFF-1 alone is sufficient for cell-cell fusion in heterologous systems24, suggesting that it drives membrane rearrangements without additional proteins. Such an independent function could explain why the standard phagosome sealing machinery is dispensable for tail-spike-cell distal-process clearance. We noted that eff-1(ns634) mutants, which contain an R77K mutation in the EFF-1 extracellular domain, are cell-fusion defective (Fig. 3h). Furthermore, EFF-1(R77K) protein localizes to plasma membrane and vesicular structures (Supplementary Fig. 2d), like wild-type EFF-1. Thus, EFF-1 fusogenic function appears to be required for phagosome sealing. Supporting this notion, neither EFF-1(R77K) nor EFF-1(G316E) (also fusion defective, and properly localized; B. Podbilewicz; pers. comm.) rescue the distal-process clearance defect of eff-1(ns634) mutants (Fig. 4h; Supplementary Fig. 2a).
If indeed EFF-1 promotes phagosome sealing through autocellular fusion, we would expect the protein to localize to the convergence site of phagosome arms. To test this, we examined localization of the EFF-1(T173A;N529D)::GFP fusion-defective protein, which shows proper localization in other contexts25,26, in an eff-1(ns634) mutant. In 14/15 animals, EFF-1 localized at the tail-spike cell remnant. Strikingly, in animals where a phagosome opening is seen, EFF-1 localizes to phagosome-arm tips (5/5; Fig. 4i–l; Supplementary Movie S15), supporting the notion that EFF-1 promotes phagosome sealing through autocellular fusion.
We demonstrate here that different morphologically-complex cell types, an epithelial cell and a neuron, undergo similar events dismantling and clearing their cell soma and processes. That two disparate C. elegans cell types are similarly eliminated raises the possibility that neurons and other morphologically complex cells are removed by related mechanisms across animals.
Different programs appear to promote degeneration of distinct cellular domains, and dismantling initiation of each domain proceeds independently of the others. Cell somas undergo morphological changes resembling apoptosis, and require canonical apoptotic engulfment genes for clearance. Proximal processes undergo caspase-dependent fragmentation and beading, reminiscent of Wallerian degeneration. However, mutations in genes required for Wallerian degeneration in Drosophila and in mice, and for LCD in C. elegans, do not block tail-spike-cell proximal-process degeneration and clearance (tir-1/Sarm or btbd-2; Supplementary Table 1;6,27), suggesting that an alternative molecular program may underlie this form of degeneration. Distal processes undergo retraction into a varicosity.
Separate clearance mechanisms also drive compartment removal, and we identify a key player, EFF-1/fusogen, required for the clearance of the distal process varicosity. Sealing of phagosomes and endocytic vesicles has been proposed to occur through the action of the protein dynamin28, or in the case of dynamin-independent endocytosis, through bar-domain proteins29. Nonetheless, whether these factors are directly responsible for membrane fusion is debated. Unlike previously-described membrane-scission factors, EFF-1 acts exoplasmically. Our studies are consistent with a model in which EFF-1 promotes auto-cellular fusion to generate a sealed phagosome in much the same way it promotes fusion of two cells (Fig. 5m, n). Such EFF-1-mediated auto-cellular fusion is also proposed in axonal regeneration30,31, dendrite sculpting14,32 and in single-cell tube formation33. EFF-1 is necessary and sufficient to drive cell fusion,34 and could similarly be the only factor required for phagosome sealing.
Recently, an EFF-1-related protein, HAP2, was found to promote gamete fusion across several phyla35. As EFF-1, HAP2, and class II fusogens share limited sequence similarity, it is possible that yet uncharacterized, but structurally-related proteins in other eukaryotes also promote phagosome sealing.
Materials and Methods
C. elegans methods
C. elegans strains were cultured using standard methods36 and were grown at 20°C. Wild-type animals were the Bristol N2 subspecies. For most tail-spike cell (TSC) experiments, one of three integrated reporters were used: nsIs435, nsIs528 or nsIs685. Integration of extrachromosomal arrays was performed using UV and trioxalen (Sigma T2137). For most experiments, animals were scored at 20°C, with the exception of the ced-3 experiment, which was done at 25°C.
Mutants
The list of mutant C. elegans strains used in this study is as follows:
LGI: ced-1(e1735), ced-1(n2089), cup-2(ar506), ced-12(ky149)
LGII: eff-1(hy21), aff-1(tm2214)
LGIII: ced-6(n2095), ced-7(n1982), tir-1(qd4)
LGIV: ced-3(n717), ced-3(n2427), ced-5(n1812), ced-2(e1752), ced-10(n1993), sand-1(or522), sand-1(ok1963), lst-4(tm2423), tag-30/btbd-2(gk474281)
LGX: ced-8(n1891), dyn-1(ky51), piki-1(ok2346)
Germline transformation and rescue experiments
Germline transformation was carried out as previously described37. All plasmids were injected at between 1–20 ng/ul. pUC19 was used to adjust the DNA concentration of injection mixtures if necessary. All rescue experiments were done with myo-2p::GFP as a co-injection marker along with cdh-3p::mcherry to label hyp10 and TSC.
Transgenes
Described in Supplementary Table 4. The full length or fragment of the aff-1 promoter was used to label the tail-spike cell. To label hyp10, either the cdh-3 promoter (embryo imaging only) or the eff-1 promoter (all other experiments) was used.
Primers and Plasmid Construction
Primer sequences and information on construction of plasmids used in this study are provided in Supplementary Table 3.
Scoring of tail-spike cell
TSC death was scored at the L1 stage. Animals were synchronized by treating gravid hermaphrodites with alkaline bleach and allowing the eggs to hatch in M9 medium overnight. Synchronized L1s were then mounted on slides on 2% agarose-water pads, anaesthetized in 10 mM sodium azide, and examined on a Zeiss Axioplan 2 or Axio-Scope A1 under Nomarski optics and wide-field fluorescence at 40x. The TSC was identified by green fluorescence (from reporter transgenes) as well as by its location and morphology.
Mutagenesis and mutant identification for the screen in this study
nsIs435 animals were mutagenized using 75 mM ethylmethanesulfonate (EMS Sigma M0880) for 4 hours at 20°C. 27,000 F2 progeny were screened for tail-spike cell persistence on a Zeiss Axio-Scope A1 at 40x. eff-1(ns634) and eff-1 (ns627) were mapped to segment 9 of Chromosome II by Hawaiian SNIP-SNP mapping38. The gene was identified by fosmid rescue and candidate gene analysis. See Supplementary Table 5.
Quantification of apoptotic cell corpse persistence
General engulfment defects were scored as persistent corpses in the heads of L1 larvae anesthetized with 30 mM sodium azide and mounted on agar pads, using Nomarski optics. n=70 for WT and 78 for eff-1(ns634).
Mosaic analysis
Three lines of eff-1(ns634); TSC::myrGFP animals with unstable extrachromosomal arrays of eff-1p::eff-1 cDNA-SL2-myr-mCherry were studied. Animals that either had mCherry (and hence eff-1) expression in hyp10, or that did not were scored for a TSC defect. Non-transgenic animals were used as controls.
Scoring cell fusion
The apical junctional marker AJM-1::GFP was used to score cell fusion defects indicated by the presence of AJM-1::GFP labeled membrane. We define fusion as the loss of AJM-1::GFP from junctions34.
Heat-shock experiments
Transgenic animals carrying a heat-inducible hsp-16.2 promoter driving eff-1 cDNA and an SL2 myristoylated mCherry were generated. These animals and siblings without the array at the synchronized L1 stage were heat-shocked in a water bath at 33°C and allowed to recover at 20°C for two hours in liquid (M9) culture. Animals were then scored for either TSC process persistence or hyp10 fusion (AJM-1::GFP) on slides.
Electron microscopy
eff-1(ns634); nsIs435 L1 larvae were imaged using a Zeiss Axioplan 2 compound microscope to measure the relative location of the TSC within the worm relative to the tail tip using the AxioVision software (Zeiss). Animals were then fixed, stained, embedded in resin, and sectioned using standard methods39. Images were acquired on an FEI TECNAI G2 Spirit BioTwin Transmission Electron Microscope with a Gatan 4K × 4K digital camera at The Rockefeller University EM Resource Center.
Microscopy and Image Processing
Some images were collected on an Axioplan 2 microscope (Zeiss) with 63x/1.4 NA objective (Zeiss) and dual-band filter set (Chroma, set 51019). Most images were collected on a DeltaVision Core imaging system at the Rockefeller Bioimaging Facility (GE Healthcare Life Sciences) with a with an Olympus IX-71 microscope and Insight 7 color SSI illumination system using an UPLSAPO 60x/1.3 NA Silicone oil objective (Olympus) and a pro.edge sCMOS camera. Images were acquired and deconvolved using measured PSFs using SoftWorx software (GE Healthcare Life Sciences). For still embryo imaging, embryos were anesthetized using 0.5 M sodium azide. Larvae were paralyzed as described above.
Fluorescence recovery after photobleaching
FRAP experiments were performed on a DeltaVision Core imaging system at the Rockefeller Bioimaging Facility using the 410, 488 and 532 laser module (QLM). Live L1 larvae were mounted and immobilized using 10 mM sodium azide. GFP surrounding the TSC was selectively photobleached at 25% 406 nm laser power for a 0.250 second pulse. Post-bleach images were collected at 5 sec intervals for 3 min. Some animals showed GFP recovery later and were imaged 30 min after photobleaching.
Scoring CEM neurons
CEM examination was done following16. Embryos were mounted using 20 mm bead spacers or 2% agarose pads. During heat-shock induction, the cell was irradiated for five continuous minutes. Precursor cells were identified using a nuclear UNC-130-GFP marker. When four cells were labeled and the signal co-localized with UNC-130-GFP, induction was scored as specific.
iSPIM imaging
Light sheet microscope imaging was performed on a custom-built inverted selective plane illumination microscope (iSPIM) based a previously published design 11 using a Hammamatsu Orca Flash 4.0 camera. Embryos collected by dissecting gravid adults were mounted on a coverslip with a polylysine solution (Sigma) spot. Imaging conditions were as follows: temperature: 20–22°C, laser power (488 nm solid state laser): 100–130 microWatts, exposure time 10.2–20 ms, time between stacks, 2–3 minutes.
Tail-spike cell ablation
Embryonic ablations were performed using an Olympus UPLSAPO 60xs objective on a Zeiss AxioObserver Z1 frame equipped with a Yokogawa CSU-X1 spinning disk head and two Hamamatsu C9100-13 EM-CCD cameras. Ablations were performed on ced-3(n717) embryos to verify successful ablation. Embryos at the 2 or 4-cell stage were cut from gravid hermaphrodites and mounted based on standard protocols40. TSC precursors were identified by tracing the embryonic lineage up until their terminal division. A total of 35 low-energy pulses were delivered at 2 Hz to each of the TSC precursors 7 minutes after observing the beginning of each cell’s cytokinesis. The ablated TSC precursors and their hyp10 siblings were then tracked to verify that the TSC precursors died and were extruded from the embryo while the hyp10 cells migrated to their correct final position and showed no signs of damage. Embryos in which one or more TSC precursors failed to be extruded or in which off-target damage was observed in neighboring cells were burst using a high-energy laser pulse. The embryos were allowed to hatch overnight before the resulting larvae were recovered and mounted for imaging.
Statistics and Reproducibility
The samples sizes and statistical tests were selected based on previous studies with similar methodologies. Sample sizes were not determined using statistical methods. All experiments were repeated at least two to three times, as indicated, giving similar results. Independent transgenic lines were treated as independent experiments. Quantification of TSC persistence was done using an unpaired, two-tailed t-test (Graphpad). For all figures, +/− SEM is represented. For Figure 4b, n=1, Supplementary Figure 1a, n=1; Supplementary Figure 1b n=2, Supplementary Figure 1f–i n=2 and Supplementary Figure g–r n=1, where n= number of biologically independent animals. Source data for Figures 1a–d, e–h, j–m-r–u, v; Figure 2a, d; Figure 3a, d; Figure 4a–g; Supplementary Figure 1a–i, Supplementary Figure 2b,c,d; Supplementary Figure 3a–i; Supplementary Figure 4a–d, g–r can be found in Supplementary Table 2.
Data Availability
Source data for Fig. 1–4 and Supplementary Fig. 2 have been provided as Supplementary Table 2. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
We thank members of the Shaham lab for discussions and critical comments on the manuscript. We thank Fabien Soulavie and Meera Sundaram for sharing data. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), and the National Bioresource Project of Japan. The work was supported by a Rockefeller Women and Science Fellowship and NIH grant 1F32HD089640 to PG; and by NIH grants R01NS081490 and R01NS078703 to SS.
Footnotes
Author contributions
PG and SS wrote the manuscript and analyzed the results. AR and PI performed iSPIM imaging. AS and AR performed CEM imaging. MT helped map mutants. PS and ZB performed tail-spike-cell ablations. YL performed EMs. PG performed all other experiments.
References and Notes
- 1.Lekstrom-Himes JA, Gallin JI. Immunodeficiency diseases caused by defects in phagocytes. The New England journal of medicine. 2000;343:1703–1714. doi: 10.1056/NEJM200012073432307. [DOI] [PubMed] [Google Scholar]
- 2.Levin R, Grinstein S, Canton J. The life cycle of phagosomes: formation, maturation, and resolution. Immunological reviews. 2016;273:156–179. doi: 10.1111/imr.12439. [DOI] [PubMed] [Google Scholar]
- 3.Mallat M, Marin-Teva JL, Cheret C. Phagocytosis in the developing CNS: more than clearing the corpses. Current opinion in neurobiology. 2005;15:101–107. doi: 10.1016/j.conb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Hochreiter-Hufford A, Ravichandran KS. Clearing the dead: apoptotic cell sensing, recognition, engulfment, and digestion. Cold Spring Harbor perspectives in biology. 2013;5:a008748. doi: 10.1101/cshperspect.a008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddien PW, Horvitz HR. The engulfment process of programmed cell death in caenorhabditis elegans. Annual review of cell and developmental biology. 2004;20:193–221. doi: 10.1146/annurev.cellbio.20.022003.114619. [DOI] [PubMed] [Google Scholar]
- 6.Osterloh JM, et al. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science. 2012;337:481–484. doi: 10.1126/science.1223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simon DJ, et al. Axon Degeneration Gated by Retrograde Activation of Somatic Pro-apoptotic Signaling. Cell. 2016;164:1031–1045. doi: 10.1016/j.cell.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohler WA, et al. The type I membrane protein EFF-1 is essential for developmental cell fusion. Developmental cell. 2002;2:355–362. doi: 10.1016/s1534-5807(02)00129-6. [DOI] [PubMed] [Google Scholar]
- 9.Chiorazzi M, et al. Related F-box proteins control cell death in Caenorhabditis elegans and human lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3943–3948. doi: 10.1073/pnas.1217271110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maurer CW, Chiorazzi M, Shaham S. Timing of the onset of a developmental cell death is controlled by transcriptional induction of the C. elegans ced-3 caspase-encoding gene. Development. 2007;134:1357–1368. doi: 10.1242/dev.02818. [DOI] [PubMed] [Google Scholar]
- 11.Wu Y, et al. Inverted selective plane illumination microscopy (iSPIM) enables coupled cell identity lineaging and neurodevelopmental imaging in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17708–17713. doi: 10.1073/pnas.1108494108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nehme R, et al. Transcriptional upregulation of both egl-1 BH3-only and ced-3 caspase is required for the death of the male-specific CEM neurons. Cell death and differentiation. 2010;17:1266–1276. doi: 10.1038/cdd.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singhal A, Shaham S. Infrared laser-induced gene expression for tracking development and function of single C. elegans embryonic neurons. Nature communications. 2017;8:14100. doi: 10.1038/ncomms14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oren-Suissa M, Hall DH, Treinin M, Shemer G, Podbilewicz B. The fusogen EFF-1 controls sculpting of mechanosensory dendrites. Science. 2010;328:1285–1288. doi: 10.1126/science.1189095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu YC, Horvitz HRC. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature. 1998;392:501–504. doi: 10.1038/33163. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- 17.Kinchen JM, Ravichandran KS. Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature. 2010;464:778–782. doi: 10.1038/nature08853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellis RE, Jacobson DM, Horvitz HR. Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics. 1991;129:79–94. doi: 10.1093/genetics/129.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koppen M, et al. Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nature cell biology. 2001;3:983–991. doi: 10.1038/ncb1101-983. [DOI] [PubMed] [Google Scholar]
- 20.Cheng S, et al. PtdIns(4,5)P(2) and PtdIns3P coordinate to regulate phagosomal sealing for apoptotic cell clearance. The Journal of cell biology. 2015;210:485–502. doi: 10.1083/jcb.201501038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo P, Hu T, Zhang J, Jiang S, Wang X. Sequential action of Caenorhabditis elegans Rab GTPases regulates phagolysosome formation during apoptotic cell degradation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18016–18021. doi: 10.1073/pnas.1008946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fares H, Greenwald I. Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics. 2001;159:133–145. doi: 10.1093/genetics/159.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumari S, Mayor S. ARF1 is directly involved in dynamin-independent endocytosis. Nature cell biology. 2008;10:30–41. doi: 10.1038/ncb1666. [DOI] [PubMed] [Google Scholar]
- 24.Podbilewicz B, et al. The C. elegans developmental fusogen EFF-1 mediates homotypic fusion in heterologous cells and in vivo. Developmental cell. 2006;11:471–481. doi: 10.1016/j.devcel.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 25.del Campo JJ, et al. Fusogenic activity of EFF-1 is regulated via dynamic localization in fusing somatic cells of C. elegans. Current biology : CB. 2005;15:413–423. doi: 10.1016/j.cub.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 26.Smurova K, Podbilewicz B. Endocytosis regulates membrane localization and function of the fusogen EFF-1. Small GTPases. 2016:1–4. doi: 10.1080/21541248.2016.1211399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinet MJ, et al. HSF-1 activates the ubiquitin proteasome system to promote non-apoptotic developmental cell death in C. elegans. eLife. 2016:5. doi: 10.7554/eLife.12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Bliek AM, Meyerowitz EM. Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature. 1991;351:411–414. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]
- 29.Simunovic M, et al. Friction Mediates Scission of Tubular Membranes Scaffolded by BAR Proteins. Cell. 2017;170:172–184 e111. doi: 10.1016/j.cell.2017.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neumann B, et al. EFF-1-mediated regenerative axonal fusion requires components of the apoptotic pathway. Nature. 2015;517:219–222. doi: 10.1038/nature14102. [DOI] [PubMed] [Google Scholar]
- 31.Ghosh-Roy A, Wu Z, Goncharov A, Jin Y, Chisholm AD. Calcium and cyclic AMP promote axonal regeneration in Caenorhabditis elegans and require DLK-1 kinase. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:3175–3183. doi: 10.1523/JNEUROSCI.5464-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oren-Suissa M, Gattegno T, Kravtsov V, Podbilewicz B. Extrinsic Repair of Injured Dendrites as a Paradigm for Regeneration by Fusion in Caenorhabditis elegans. Genetics. 2017;206:215–230. doi: 10.1534/genetics.116.196386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasmussen JP, English K, Tenlen JR, Priess JR. Notch signaling and morphogenesis of single-cell tubes in the C. elegans digestive tract. Developmental cell. 2008;14:559–569. doi: 10.1016/j.devcel.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shemer G, et al. EFF-1 is sufficient to initiate and execute tissue-specific cell fusion in C. elegans. Current biology : CB. 2004;14:1587–1591. doi: 10.1016/j.cub.2004.07.059. [DOI] [PubMed] [Google Scholar]
- 35.Fedry J, et al. The Ancient Gamete Fusogen HAP2 Is a Eukaryotic Class II Fusion Protein. Cell. 2017;168:904–915 e910. doi: 10.1016/j.cell.2017.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. The EMBO journal. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nature genetics. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- 39.Lundquist EA, Reddien PW, Hartwieg E, Horvitz HR, Bargmann CI. Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development. 2001;128:4475–4488. doi: 10.1242/dev.128.22.4475. [DOI] [PubMed] [Google Scholar]
- 40.Bao Z, Murray JI. Mounting Caenorhabditis elegans embryos for live imaging of embryogenesis. Cold Spring Harbor protocols. 2011;2011 doi: 10.1101/pdb.prot065599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for Fig. 1–4 and Supplementary Fig. 2 have been provided as Supplementary Table 2. All other data supporting the findings of this study are available from the corresponding author on reasonable request.