Abstract
Repair of complex CNS circuitry will require newly incorporated neurons to become appropriately, functionally integrated. One approach is to direct differentiation of endogenous progenitors in situ, or ex vivo followed by transplantation. Prior studies find that newly incorporated neurons can establish long-distance axon projections, form synapses, and functionally integrate in evolutionarily old hypothalamic energy balance circuitry. We now demonstrate that postnatal neocortical connectivity can be reconstituted with point-to-point precision, including cellular integration of specific, molecularly identified projection neuron subtypes into correct position, combined with development of appropriate long-distance projections and synapses. Using optogenetics-based electrophysiology, experiments demonstrate functional afferent and efferent integration of transplanted neurons into trans-callosal projection neuron circuitry. Results further indicate that “primed” early postmitotic neurons, including already fate-restricted deep layer projection neurons and/or plastic post-mitotic neuroblasts with partially fate-restricted potential, account for the predominant population of neurons capable of achieving this optimal level of integration.
Introduction
A long-standing question in nervous system regenerative biology is whether immature progenitors or pluripotent cells can be directed to even partially regenerate circuits affected in neurodegenerative and acquired disorders. Behaviorally relevant repair of circuitry in the central nervous system (CNS) will require that at least some newly incorporated neurons become appropriately and electrophysiologically integrated. One developmentally-motivated approach is to direct differentiation of endogenous progenitors within the brain itself, or progenitors/pluripotent stem cells ex vivo, followed by transplantation. A critical brain region for regeneration is cerebral cortex, disrupted in a variety of human disorders. Even modest re-formation of circuitry, if relatively specific, might enable functionally meaningful restoration of lost connectivity.
Work from our lab demonstrated that evolutionarily old and conserved, homeostatic circuitry of hypothalamus, with diverse neuron populations organized into nuclei, can be anatomically, molecularly, and functionally restored. Micro-transplanted immature neurons differentiated into appropriate subtypes and restored physiological energy balance signaling, with organism-level behavioral rescue1. Work from other labs demonstrates that transplanted interneurons are capable of migrating and integrating into recipient circuitry2–10.
Previous work from our lab and others demonstrated that transplant-derived immature cortical projection neurons, or neurons newly recruited in situ, can establish long-distance anatomic connectivity in the rodent CNS11–17. Several studies report establishment of long-distance axon projections, some with formation of synapses and/or afferent integration18–23. It remains unknown, however, whether long-distance cortical projection neuron connectivity can be regenerated with point-to-point specificity of projection neuron subtype diversity.
Effective integration of newly incorporated cortical neurons would require multi-step circuit formation: differentiation into neurons with defined, mature subtype identities, without subtype “confusion”; cellular-anatomic integration into correct laminar positions to receive appropriate inputs; and establishment of appropriate, functional output connectivity. We designed experiments to investigate these cardinal features of projection neuron integration and specificity.
We performed micro-transplantation of eGFP+ embryonic neurons, encompassing already fate-restricted deep layer projection neurons and/or still plastic post-mitotic neuroblasts with partially fate-restricted potential (“primed” for a relatively narrow range of differentiation), into early-postnatal somatosensory cortex, and employed newly available markers of neocortical projection neuron subtype-specificity24,25, plus optogenetics-based slice electrophysiology.
Results demonstrate that developmentally primed neurons build long-distance inter-hemispheric (trans-callosal) and subcerebral connectivity with cellular integration of molecularly identified projection neuron subtypes into correct laminar position, with development of appropriate long-distance axonal projections. Remarkably, the concordance of established subcerebral or trans-callosal connectivity and molecular identity of transplanted neurons was 100%.
These results have implications both toward repair of complex CNS circuitry, and for directed differentiation from embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs) for in vitro investigation and therapeutic screening, for which precise differentiation into distinct projection neuron subtypes is often critical.
Results
To most deeply and rigorously investigate these issues at the single-neuron level, thus enabling investigation with multiple molecular identity, connectivity, birthdate, and electrophysiologic analyses on individual identified neurons, we investigated a relatively small number of neurons with each combination of analyses. We investigated a priori only transplanted neurons that had integrated within cortical laminae, and were separated from cortical white matter by at least one layer of recipient neurons. Micro-transplantation was optimized for sparse neuronal integration, enabling the desired single neuron resolution analysis. Since multiple aspects of neuronal identity are interrogated via molecular, birthdate, and/or connectivity analyses, overall numbers are n=9–13 for integrated analysis of a specific neuronal subtype identity and its fidelity of connectivity, while more than n=3 neurons were typically investigated with specific combinations of molecular markers, connectivity, and birthdating. Fisher’s exact test enabled statistical analysis of molecular-to-projection fidelity. Optogenetics-based, patch-clamp-electrophysiology enabled investigation of afferent and efferent integration of transplanted neurons into trans-callosal projection neuron circuitry, again with relatively small numbers for depth of analysis of individual identified neurons. For example, 26 recipient-derived neurons were recorded, 2 ipsilateral and 24 contralateral to transplantation sites. Detailed sample size information is presented in Methods and in Figures.
Developmentally primed immature neurons integrate as distinct molecular projection neuron subtypes
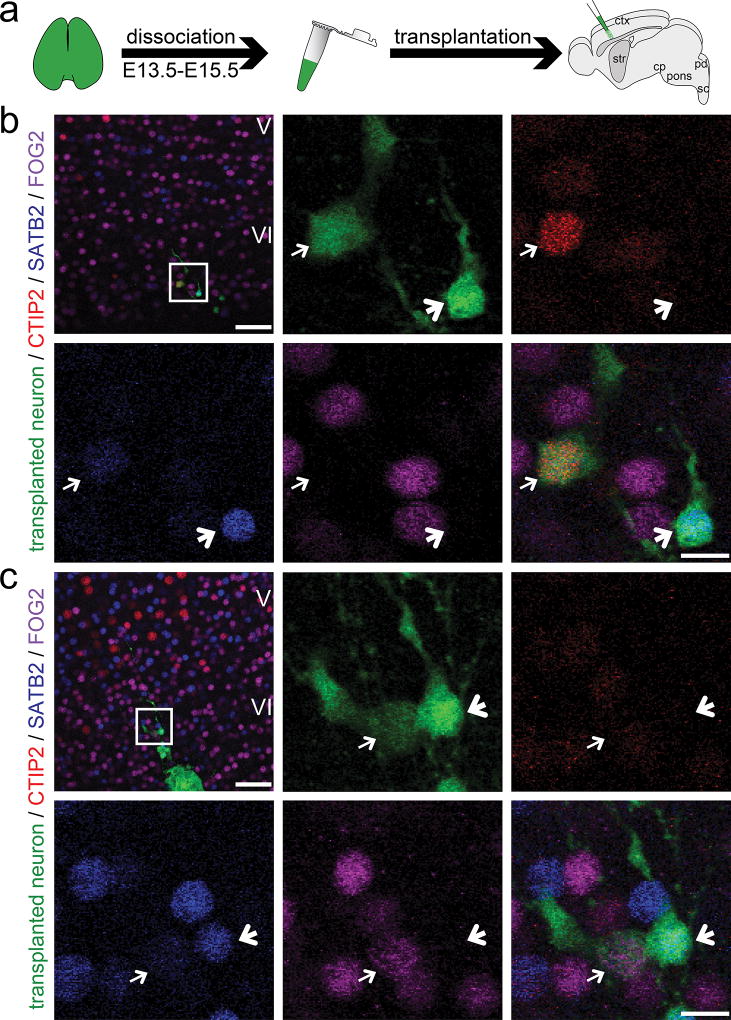
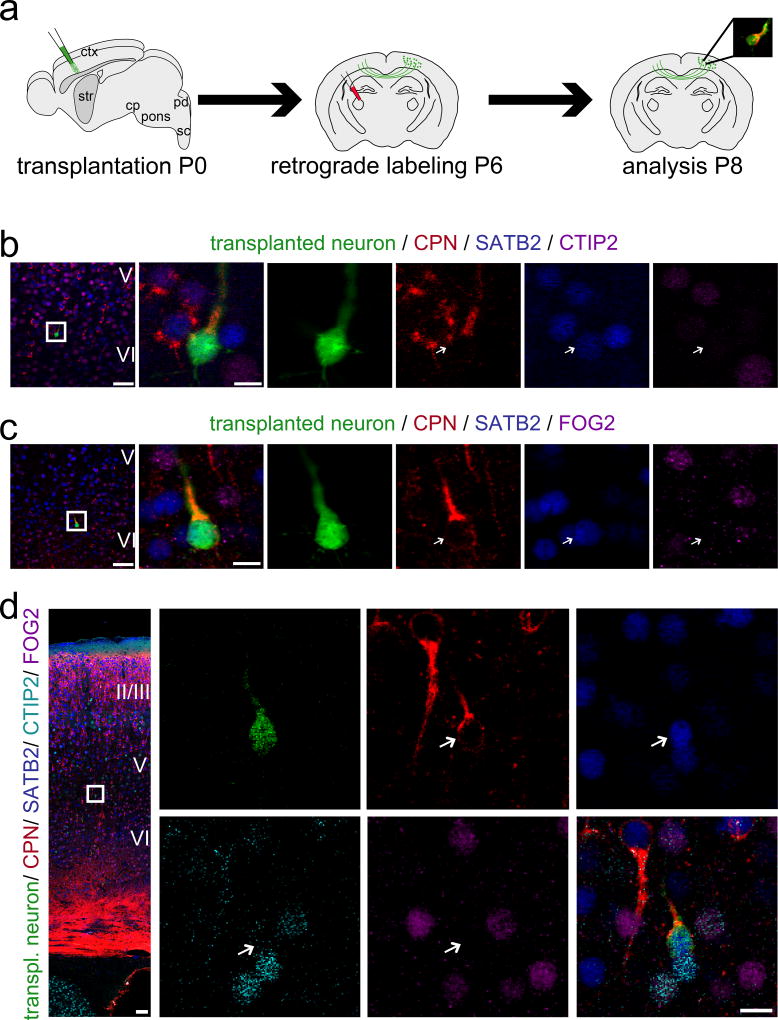
To determine whether developmentally primed immature neurons retain ability to differentiate into distinct molecular projection neuron subtypes following transplantation into postnatal neocortical circuitry (Fig. 1a), we evaluated combinatorial expression of recently identified transcriptional controls CTIP226, SATB227,28, and FOG224,29. During postnatal development, corticospinal/subcerebral projection neurons (CSMN/SCPN) are characterized by high-level expression of transcriptional controls CTIP2 (and Fezf230), while negative for SATB2 and FOG2. Callosal projection neurons (CPN) express SATB2, while negative for CTIP2 (and Fezf2) and FOG2. Corticothalamic projection neurons (CThPN) express FOG2, and both CTIP2 and Fezf2 at low levels, while negative for SATB224,25.
Figure 1. Developmentally primed immature neurons integrate as distinct molecular projection neuron subtypes.
(a) Sensorimotor cortices of eGFP+ donor embryos between E13.5 and E15.5 are dissociated into a single cell suspension. Donor neurons are then micro-transplanted into early postnatal recipient sensorimotor cortex and analysis is performed at P6. (b, c) Transplanted eGFP+ neurons integrate as distinct molecular projection neuron subtypes: (b) SCPN (thin arrows: CTIP2 high-level, SATB2 very low-level, FOG2 negative), (b, c) CPN (bold arrows: CTIP2 negative, SATB2 positive, FOG2 negative), (c) CThPN (thin arrows: CTIP2 negative/low-level, SATB2 very low-level, FOG2 positive). The positions of the boxes in low magnification panels (b, c) correspond to the cortical area examined in high magnification panels. eGFP (green); CTIP2 (red); SATB2 (blue); FOG2 (purple). Scale bars: b and c (low magnification), 50 µm; b and c (high magnification), 10 µm.
Immunocytochemistry reveals that a subset of transplant-derived, developmentally primed neurons that migrate away from transplantation tracks and are isolated from heterotopic clusters indeed maintain and/or develop distinct molecular identity of either SCPN (Fig. 1b; thin arrows), CPN (Fig. 1b, c; bold arrows), or CThPN (Fig. 1c; thin arrows). They do not assume mixed identity, nor a “generic” and less specific projection neuron phenotype. Together, these molecular marker data suggest that at least some transplanted neurons continue differentiation with fidelity to integrate as distinct molecular neuronal subtypes within recipient cortex.
Transplanted neurons assuming positions in cortical layers II–VI progressively mature and successfully extend long-distance axon projections
Confirming earlier work of our group and others, many newly incorporated neurons establish long-distance projections, either through the internal capsule to subcortical locations (Fig. S1a – e and f) or across the corpus callosum to contralateral cortex (Fig. S1h – j and g). Axons pass through the internal capsule (Fig. S1a) and extend progressively, reaching the cerebral peduncle (cp) by 4 days post-transplantation (dpt) (Fig. S1b), distal pons (Fig. S1c) and proximal medulla (Fig. S1d) by 5 dpt, and distal medulla by 6 dpt (Fig. S1e), reaching the contralateral dorsal funiculus of the cervical spinal cord by 12 dpt (data not shown). Similarly, trans-callosal axons reach contralateral cortex by 5–6 dpt (Fig. S1h – j). Recent work by others demonstrates that human and mouse embryonic stem cell (hES; mES)-derived neurons transplanted into early postnatal or lesioned adult brains are capable of projecting axons to trans-callosal and corticofugal targets, though often quite multidirectionally, along existing axon tracts. Most hES/mES-derived neurons in these studies form heterotopic aggregates spanning cortical gray and white matter, with only rare neurons observed to disperse into surrounding cortex. Retrograde labeling from corticofugal targets in these studies revealed that axons originated from the predominant population of neurons within non-integrated heterotopias, not from rare, more cellularly integrated neurons21,22.
To investigate whether such limitations can be overcome, and whether integration into cortical circuitry can be further refined, we performed micro-transplantation (details in Methods) of small numbers of developmentally primed immature neurons using established methods1,13,14.
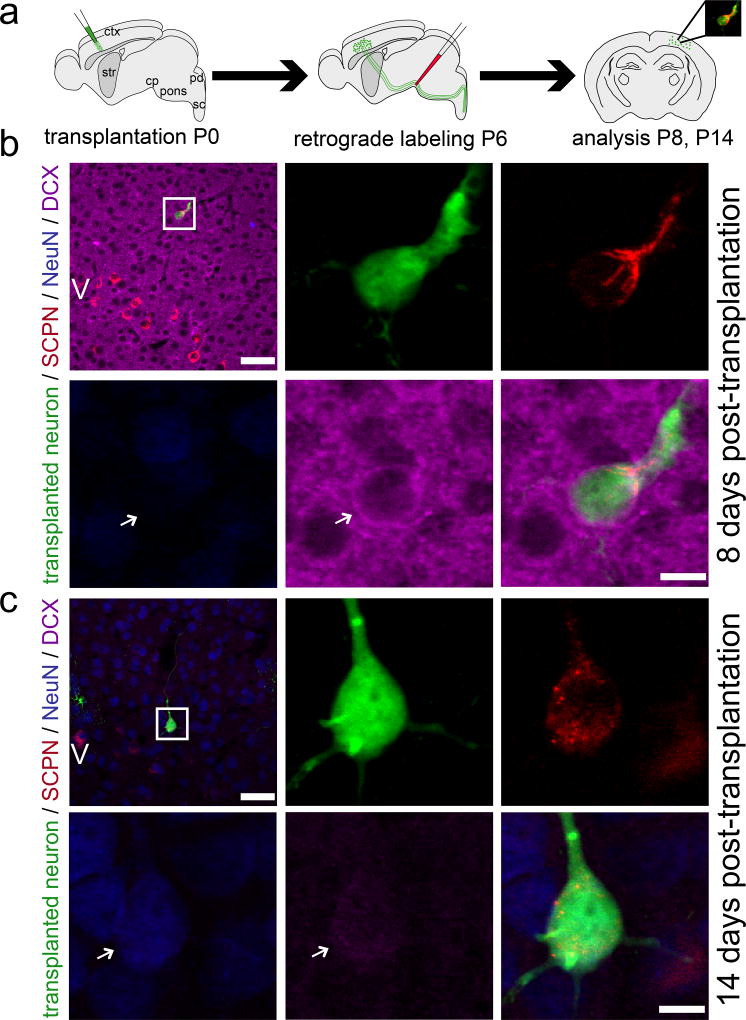
Retrograde labeling with CTB555 from the cerebral peduncle 5–6 dpt (Fig. 2a), or from contralateral cortex 5–6 dpt (Fig. S2a) and 35 dpt (Fig. S3a) reveals unambiguously that a subset of neurons assume positions as integrated single neurons in cortical layers II–VI (not in white matter tracts or heterotopic aggregates), and successfully establish long-distance subcerebral or trans-callosal projections. They continue to mature in vivo over 2–5 weeks from an initially DCX+ state (immature, migratory neuroblast stage) to a DCX−/low and NeuN+ state (mature neuronal stage), closely paralleling the maturational time course of surrounding recipient/endogenous cortical neurons (SCPN: Fig. 2b, c; CPN Fig. S2b, c). Importantly, axons of these neurons remain stable long-term, without pruning or retraction (as long as analyzed; 5 weeks) (CPN: Fig. S3b).
Figure 2. Transplanted neurons assuming positions in cortical layers II–VI progressively mature and successfully extend long-distance subcerebral axon projections.
(a) Schematic of micro-transplantation of eGFP+ single cell suspension into P0/P1 recipient sensorimotor cortex. Ultrasound-guided injection of retrograde axonal tracer (CTB555) into the cerebral peduncle (cp) at P6. Analysis at P8 and P14. (b, c) A subset of transplanted neurons project subcerebrally. These neurons progressively mature from being DCX+ and NeuN− (b) to being DCX−/low and NeuN+ (c). Comparison with the DCX expression of the recipient cortex at both time points reveals that DCX of the recipient cortex decreases dramatically and on a very similar time scale to that of the transplanted neurons. The positions of the boxes in low magnification panels (b, c) correspond to the cortical area examined in high magnification panels. Arrows indicate transplanted neurons. eGFP (green), CTB555 (red), DCX (immature neuronal marker; purple), NeuN (mature neuronal marker; blue). Scale bars: b and c (low magnification), 50 µm; b and c (high magnification), 10 µm.
Double birthdating donor neurons before transplantation to investigate stage and origin of cells mediating successful cortical integration
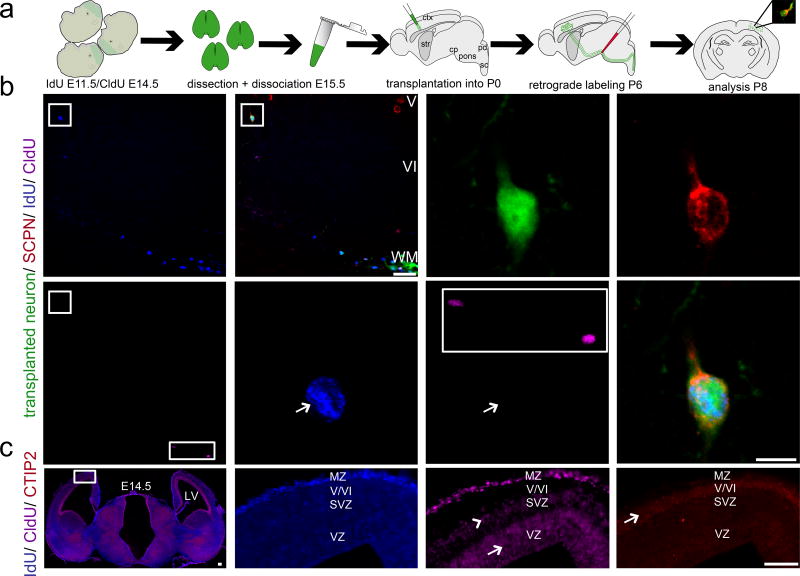
To investigate the stage and origin of cells contributing to successful long-distance cortical integration of transplant-derived projection neurons, transplanted neurons were first double birthdated in utero using IdU (E11.5 onward) and CldU (E14.5 onward, 16 hours before dissociation) within donor embryos. Then, transplanted neurons were retrogradely labeled from the cerebral peduncle, and individual neurons analyzed for connectivity and developmental stage/birthdate (Fig. 3a). Retrogradely labeled neurons were postmitotic >16 hours before transplantation (IdU+ / CldU−, Fig. 3b). Some transplanted (but not retrogradely labeled) neurons within the same sections were CldU+, serving as internal positive controls for CldU immunocytochemistry (Fig. 3b, rectangular boxes). Three additional examples are presented in Fig. S4. Analysis of donor littermate cortex in vivo identified that corresponding postmitotic neurons at this gestational stage had already formed deep cortical layers, already CTIP2+ (Fig. 3c). As expected, all neurons in cortical plate were IdU+, with CldU+ mitotic cells in the VZ (arrow) and both dividing progenitors and early postmitotic neuroblasts in the SVZ (arrowhead), mainly destined to generate superficial layers II/III. Peak neurogenesis for layers V–VI was largely complete by E14.5, the time of CldU injection (layer V [arrow] with strong CTIP2 expression; Fig. 3c). Thus, cells in layers V–VI were CldU− (Fig. 3c). These data indicate that early postmitotic DCX+ neuroblasts destined to form deep layer projection neurons, that had already assumed appropriate positions in cortical plate when dissociated, account for the predominant population of neurons integrating into recipient cortex, and establishing subcerebral projections after transplantation.
Figure 3. Double birthdating of donor neurons prior to transplantation to investigate the stage and origin of cells mediating successful cortical integration.
(a) Schematic illustrating the experimental outline. Double birthdating of eGFP+ donor embryos in utero using IdU (from E11.5 onward) and CldU (from E14.5 onward, 16 hours prior to harvesting and dissociation), followed by dissection/dissociation of cortices at E15.5 and micro-transplantation of eGFP+ and double birthdated single cell suspension into P0/P1 recipients. Ultrasound-guided injection of retrograde axonal tracer (CTB555) into the cerebral peduncle (cp) at P6. Analysis at P8. (b) Recipient cortex after transplantation and retrograde labeling from the cp. The eGFP+ transplanted neuron (boxed area in left upper corner) and recipient SCPN are backlabeled with CTB555 from the cp. Furthermore there are two additional eGFP+/birthdated transplanted but CTB555 negative cells in the same field of view (right lower corner in (rectangular box). Both cells were found to be double-positive for IdU and CldU demonstrating that both the birthdating and the ICC were successful and indicating that these 2 cells were likely to be still mitotic or only very early postmitotic (for less than 16 hours) at the time of transplantation. Analysis of the eGFP+ and CTB555+ transplanted neuron shows that it is positive for IdU but not CldU (arrows), demonstrating that this neuron was postmitotic for more than 16 hours before transplantation. The positions of the squared boxes in low magnification panels (b) correspond to the cortical area examined in high magnification panels. The area corresponding to the rectangular box in the low power panel is shown as a magnified inset in the high magnification CldU panel for comparison. eGFP (green), CTB555 (red), IdU (blue), CldU (purple).
(c). Analysis of donor littermate cortex in vivo identified that corresponding postmitotic neurons at this gestational stage had already formed deep cortical layers, and were already CTIP2+. The left panel shows an epifluorescence montage (merge of the blue (IdU) and purple (CldU) channel) of donor littermate cortex. The positions of the box correspond to the cortical area examined in the high magnification panels (IdU, CldU). A corresponding area of an adjacent section is magnified in the CTIP2 panel (right lower corner). ICC for IdU, CldU and CTIP2 reveals the expected labeling pattern confirming that birthdating was successfully performed. As expected, all neurons in the cortical plate were labeled by IdU (administration of IdU from E11.5 on up to the dissociation time point). CldU incorporated into mitotic cells in the VZ (indicated by arrow) and a mix of dividing progenitors and early postmitotic neuroblasts in the SVZ (indicated by arrowhead), mainly destined to generate superficial layers II/III. Immunohistochemistry for CTIP2 was performed in an adjacent brain section. The peak time of neurogenesis for layers V and VI was largely complete by E14.5, the time of CldU injection (with layer V [indicated by arrow] clearly outlined by strong CTIP2 expression). Thus, cells in layers V and VI were CldU negative. CTIP2 (red), IdU (blue), CldU (purple). Scale bars: b (low magnification), 50 µm; b (high magnification), 10 µm; c, 500 µm. Double birthdating data are presented for one transplanted and retrogradely labeled SCPN; primary data from three additional neurons are presented in Fig. S4. All analyzed neurons were IdU+/CldU−. These data indicate that early postmitotic neurons, whether already fate-restricted deep layer projection neurons and/or plastic post-mitotic neuroblasts with partially fate-restricted potential, account for the predominant population of neurons capable of achieving optimal level integration, and establishing subcerebral axon projections after transplantation.
Neurons of specific combinatorial molecular identity rebuild long-distance efferent-output circuitry with high fidelity
We next investigated whether molecular identity and axonal connectivity of integrated neurons matched with fidelity, or whether they might have mixed and/or “confused” molecular identity, potentially establishing promiscuous long-distance projections. We employed combinations of positive subtype identifiers and alternative, negative, exclusionary markers (e.g. high-level CTIP2 and Fezf2, expressed by SCPN; SATB2, expressed by CPN; FOG2, expressed by CThPN). This enabled investigation of both specificity and potential “confusion” combinatorially at single cell level, simultaneously with investigation of projections by the same individual neurons (via retrograde labeling).
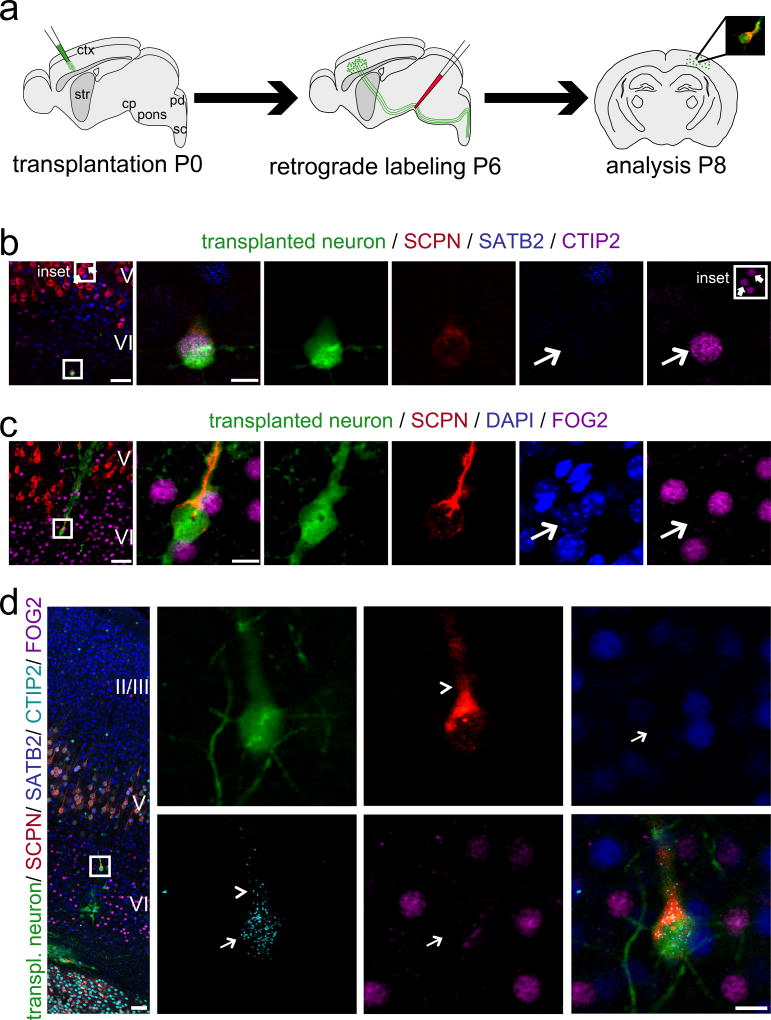
Combinatorial ICC analyses of expression of CTIP2 and SATB2 in single newly incorporated eGFP+ neurons, retrogradely labeled from cp (Fig. 4a), reveals specific expression of the SCPN marker CTIP2, and lack of the alternative callosal marker SATB2 (Fig. 4b; for additional examples Fig. S5a, c). Importantly, ICC analysis identifies high-level expression of CTIP2 comparable to that observed in endogenous layer V SCPN, rather than much lower level expression displayed by CThPN (Fig. 4b; Fig. S5a; Fig. S6). These data indicate specific SCPN, rather than broad corticofugal projection neuron (CFuPN), identity.
Figure 4. Newly incorporated neurons of combinatorial subcerebral molecular identity establish anatomical subcerebral output connectivity with high fidelity.
(a) Micro-transplantation of eGFP+ single cell suspension into P0/P1 recipient sensorimotor cortex. Ultrasound-guided injection of retrograde axonal tracer (CTB555) into the cerebral peduncle (cp) at P6. Analysis at P8. (b, c) 4-channel confocal analysis. (d) Five-channel/spectral unmixing confocal analysis. eGFP+ and subcerebrally projecting transplanted neurons express the SCPN molecular control and marker CTIP2 at high levels, and are negative for the callosal molecular control and marker SATB2 (b, d) and for the corticothalamic molecular control and marker FOG2 (c, d). Thin arrows indicate nuclei of transplanted neurons. The inset in (b) shows representative high-level CTIP2 staining of two endogenous layer V subcerebral projection neurons (block arrows) demonstrating comparable high-level CTIP2 expression by the transplanted neuron. The arrowhead in (d) points out incomplete spectral unmixing of A-594 (cyan) and A-555 (red) resulting in minor bleed through of the cytoplasmic CTB555 signal. The positions of the boxes in low magnification panels (b – d) correspond to the cortical area examined in high magnification panels. (b): eGFP (green), CTB555 (red), SATB2 (blue), CTIP2 (purple); (c): eGFP (green), CTB555 (red), DAPI (blue), FOG2 (purple); (d): eGFP (green), CTB555 (red), SATB2 (blue), CTIP2 (cyan), FOG2 (purple). Scale bars: b, – d (low magnification), 50 µm; b – d (high magnification), 10 µm. Molecular identity data are presented for three transplanted and retrogradely labeled SCPN; primary data from ten additional neurons are presented in Fig. 6; Figs. S5, 6, 8, and 12. Taken together, these data reveal a remarkable level of molecular-to-projection fidelity, even compared with the very closely related corticofugal CThPN, since neither SATB2+ nor FOG2+ transplanted neurons projected subcerebrally.
To further investigate neuronal subtype identity of these neurons, we assessed expression of FOG2, a molecular identifier of CThPN, the predominant alternative corticofugal population. Both SCPN and CThPN axons descend ~orthogonally across corpus callosal white matter into the internal capsule, then follow distinct trajectories to brain stem/spinal cord or thalamic nuclei, respectively. Our experiments reveal that only FOG2− transplanted neurons project to subcerebral targets (Fig. 4c; for additional examples Fig. S5b, c).
To confirm these data at single cell level, we combined ICC analysis and five-channel confocal imaging with spectral unmixing. This demonstrates specific expression of CTIP2, and absence of both SATB2 and FOG2 (Fig. 4d). Together, these data reveal a remarkable level of molecular to projection fidelity, even compared with very closely related CThPNs, since neither SATB2+ nor FOG2+ transplanted neurons projected subcerebrally.
To investigate both the complement of these experiments– whether callosally projecting transplanted neurons remain molecularly subtype specific– and the possibility that the sheer size of the corpus callosum might aberrantly “capture” axons of some molecularly-identified CFuPN, resulting in projection promiscuity, we transplanted developmentally primed neurons, then retrogradely labeled from contralateral cortex (Fig. 5a). We found that transplanted and endogenous neurons projecting across the callosum express comparable levels of SATB2, and are CTIP2−/low (Fig. 5b; for additional examples Fig. S7a, c). Further, transplant-derived CPN (consistent with results with transplanted SCPN) do not express FOG2 (Fig. 5c; for additional examples Fig. S7b, c). We similarly investigated callosally projecting transplanted neurons at single cell level by five-channel confocal imaging with spectral unmixing (Fig. 5d), revealing that CPN express SATB2 but not CTIP2 or FOG2. Together, all analyzed transplanted and trans-callosally projecting neurons possess CPN molecular identity, and no transplanted neurons of SCPN or CThPN molecular identity were retrogradely labeled from contralateral cortex.
Figure 5. Newly incorporated neurons of combinatorial callosal molecular identity establish anatomic inter-hemispheric output connectivity with high fidelity.
(a) Micro-transplantation of eGFP+ single cell suspension into P0/P1 recipient sensorimotor cortex. Stereotaxic injection of retrograde axonal tracer (CTB555) into the contralateral hemisphere at P6. Analysis at P8. (b, c) 4-channel confocal analysis. (d) Five-channel/spectral unmixing confocal analysis. eGFP+ and trans-callosally projecting transplanted neurons express the callosal marker SATB2 and are negative for the subcerebral marker CTIP2 (b, d) and for the corticothalamic marker FOG2 (c, d). Arrows indicate transplanted neurons. The positions of the boxes in low magnification panels (b – d) correspond to the cortical area examined in high magnification panels. (b): eGFP (green), CTB555 (red), SATB2 (blue), CTIP2 (purple); (c): eGFP (green), CTB555 (red), SATB2 (blue), FOG2 (purple); (d): eGFP (green), CTB555 (red), SATB2 (blue), CTIP2 (cyan), FOG2 (purple). Scale bars: b – d (low magnification), 50 µm; b – d (high magnification), 10 µm. Molecular identity data are presented for three transplanted and retrogradely labeled CPN; primary data from six additional neurons are presented in Fig. S7. Taken together, all analyzed transplanted and trans-callosally projecting neurons possess CPN molecular identity, and no transplanted neurons of SCPN or CThPN molecular identity were retrogradely labeled from the contralateral hemisphere.
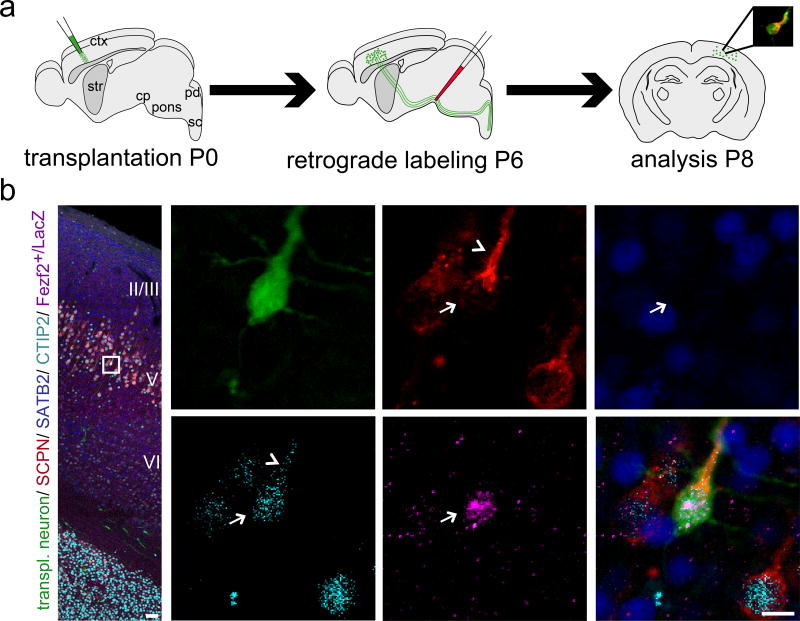
We extended molecular analysis to include Fezf2, and investigated whether SCPN (Fezf2+, CTIP2+, SATB2−) can integrate into correct laminar positions (cortical layer V) while establishing appropriate subcerebral projections.
In Fezf2+/LacZ heterozygous mice, b-gal is a reporter for Fezf2 expression, identifying Fezf2+ neurons (no specific anti-Fezf2 antibody for tissue). We crossed eGFP mice with Fezf2+/LacZ mice, yielding eGFP+/Fezf2+/LacZ donor embryonic neurons (Fig. 6a); all transplant-derived neurons were eGFP+, while additional b-gal+ ICC reported Fezf2 expression. We observed that transplant-derived and subcerebrally projecting neurons (by retrograde labeling) both have the ability to integrate within appropriate cortical layer V, and exhibit specific SCPN identity (Fezf2+, high-level CTIP2+, SATB2−) (Fig. 6b).
Figure 6. Newly incorporated neurons establish point-to-point long-distance connectivity.
(a) Micro-transplantation of eGFP+/Fezf2+/LacZ single cell suspension into P0/P1 recipient sensorimotor cortex. Ultrasound-guided injection of retrograde axonal tracer (CTB555) into the cerebral peduncle (cp) at P6. Analysis at P8. (b) Five-channel/spectral unmixing confocal analysis. The position of the box in the low magnification panel of the confocal montage corresponds to the cortical area examined in high magnification panels.. The arrow points out a transplanted and retrogradely labeled subcerebrally projecting neuron. Strikingly, this neuron integrated in anatomic correct position within layer V (surrounded by numerous recipient-derived CTB555-labeled SCPN). ICC analysis confirms SCPN molecular identity of the transplanted neuron, as revealed by Fezf2 (reported by b-gal staining) and CTIP2 expression as well as lack of expression of the exclusionary marker SATB2. The arrowhead indicates bleed-through of dense cytoplasmic CTB555 labeling of the apical dendrite when spectrally unmixing from A-594 (nuclear CTIP2 staining). Note that surrounding wild-type recipient-derived SCPN are negative for Fezf2-b-gal demonstrating the high specificity of the anti-b-gal antibody. eGFP (green), CTB555 (red), CTIP2 (cyan), SATB2 (blue), Fezf2-b-gal (purple). Scale bars: b (low magnification), 50 µm; b (high magnification), 10 µm. Molecular identity data are presented for one transplanted and retrogradely labeled SCPN that integrated into appropriate cortical layer V; primary data regarding molecular identity from twelve additional subcerebrally projecting neurons are presented in Fig. 4; Figs. S5, 6, 8, and 12. Taken together, these data reveal that transplant-derived and subcerebrally projecting neurons (by retrograde labeling) both have the ability to integrate within appropriate cortical layer V, and exhibit specific SCPN identity.
Extending from these data, demonstrating establishment of specific output connectivity, we asked whether neurons with specific combinatorial molecular identity could repopulate circuitry lacking subtype-specific neurons. Fezf2 is a transcriptional regulator required for birth and specification of SCPN30,31. In Fezf2−/− mice, SCPN are not specified, resulting in complete absence of subcerebral axon projections, thus of the corticospinal / pyramidal tract26,30,31. In a proof-of-concept experiment, we transplanted developmentally primed immature neurons into Fezf2−/− recipient cortex (Fig. S8a). Retrograde labeling from cerebral peduncle, and subsequent ICC analysis for CTIP2 and SATB2, confirmed absence of endogenous SCPN (neither high-level CTIP2 nor retrograde labeled neurons present in layer V). As expected, many medium spiny neurons (MSN) in striatum were retrogradely labeled (asterisk) due to anatomical proximity of cerebral peduncle and substantia nigra (MSN projection target) (Fig. S8b). In contrast, wt mice demonstrate high-level CTIP2 labeling and retrograde labeling of layer V SCPN, with retrograde labeling of striatal MSN (Fig. S8b, inset in right corner of left panel, asterisk). MSN labeling is an internal control, confirming correct ultrasound-guided targeting of CTB555 retrograde label into cerebral peduncle of Fezf2−/− recipient mice. Quite strikingly, newly incorporated (transplanted) neurons of SCPN molecular subtype identity (high-level CTIP2+, SATB2−) extend subcerebral projections even in the absence of the endogenous pyramidal tract, thereby reconstituting otherwise entirely absent long-distance connectivity in recipient brain (Fig. S8b, c).
These results so far reveal that, even with hundreds of thousands of intervening, aligned orthogonal projections, newly incorporated and molecularly identified SCPN do not send axons across the callosum to contralateral cortex. Rather, they maintain fidelity in establishment of long-distance connectivity. Conversely, all neurons retrogradely labeled from contralateral cortex possess CPN identity. We next investigated whether electrophysiologically functional afferent and efferent cortical projection neuron circuitry can be rebuilt.
Newly incorporated neurons functionally integrate into complex neocortical circuitry, establishing functional afferent and efferent local and long-distance synaptic connections with recipient brain
Synapse formation
Effective regeneration of circuitry will critically depend on the extent of functional synaptic restoration of connectivity. We first investigated morphological and molecular bases of afferent electrophysiological connectivity: dendritic and synapse formation. Confocal reconstructions of an eGFP+ transplanted neuron (Fig. S9a) demonstrate that soma and dendrites are tightly surrounded by recipient-derived synaptophysin+ presynaptic terminals. An epifluorescence image of the same transplanted neuron is shown in (Fig. S9b, left panel). The boxed areas are magnified on the right and depict high-resolution, high-magnification 3D confocal reconstructions of the corresponding eGFP+ apical dendrite. Close proximity of recipient-derived and transplant-derived pre- and postsynaptic elements are revealed, respectively (Fig. S9b, middle panel). This close apposition of recipient-derived presynaptic terminals and eGFP+ transplanted neuron spines is further revealed in single optical confocal sections, strongly indicating structural synaptic integration of transplanted neurons into recipient neocortical circuitry (Fig. S9b, right panels).
Channelrhodopsin-2-based mapping of afferent synaptic integration of transplanted neurons newly incorporated into long-distance trans-callosal circuitry
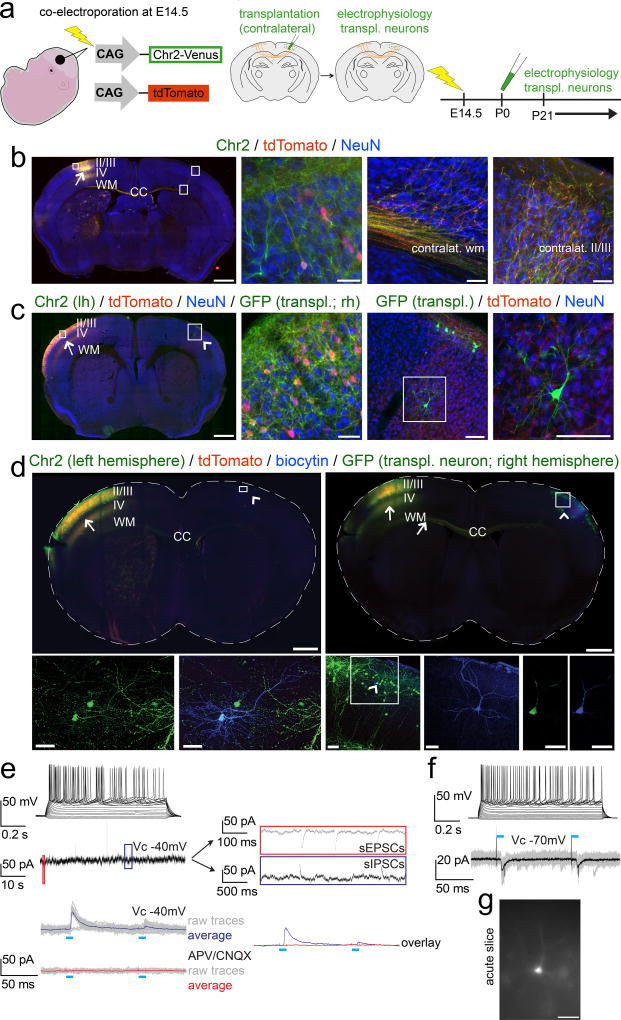
To investigate whether transplanted immature neurons can undergo functional afferent integration into local and long-distance circuitry, we developed a strategy by which a discrete number of callosally projecting recipient-derived neurons of defined laminar position (by birth-date) can be optogenetically activated to investigate their electrophysiological connectivity as afferent inputs to contralateral transplanted neurons. We used in utero-electroporation (IUE) of channelrhodopsin-2-Venus (ChR2-Venus) along with a tdTomato reporter construct into developing left cortical VZ at E14.5, the peak birth-date of layers II/III CPN, prior to postnatal contralateral transplantation of neurons in homotopic right cortex (Fig. 7a). ICC amplification of ChR2-Venus demonstrates presence of ChR2-Venus even in very distal axonal terminals of electroporated recipient (endogenous) CPN (Fig. 7b). tdTomato native fluorescence enabled transcranial visualization of the electroporated cortical area on the day of transplantation (P0/P1), enabling specific contralateral homotopic micro-transplantation. tdTomato fluorescence also visualized recipient-derived axon terminals in acute slices without activating ChR2, and guided choice of transplanted neurons for electrophysiological evaluation. Transplanted neurons surrounded by recipient-derived tdTomato+ and presumptive ChR2-Venus+ axon terminals (Fig. 7c) were selected for electrophysiological recordings to optimize efficiency of analysis of potential synaptic input from contralateral recipient CPN.
Figure 7. Newly incorporated neurons afferently integrate into long-distance trans-callosal circuitry.
(a) Schematic illustrating the experimental outline: in utero-electroporation of channelrhodopsin-2-Venus and tdTomato constructs into the left developing cortex at E14.5, targeting layer II/III CPN (left panel). Transplantation of eGFP+ neurons into the homotopic area of the contralateral cortex (identified by tdTomato fluorescence of CPN axon terminals) at P0/P1 (middle panel). Electrophysiology from 3 weeks of age on (right panel). (b) Epifluorescence montage of a coronal section after in utero-electroporation of layer II/III CPN with channelrhodopsin-2-Venus and tdTomato in the left hemisphere (arrow). The ipsilateral cortical area examined (box in left hemisphere) shows an electroporated neuron demonstrating membrane localization of channelrhodopsin-2-Venus in comparison to the cytoplasmic localization of tdTomato (high magnification panel 2). The boxed areas in the contralateral cortex, display the contralateral white and cortical gray matter, demonstrating that channelrhodopsin-2-Venus is also transported and localized to distal axonal compartments of in utero-electroporated CPN (high magnification panels 3 and 4). Immunocytochemistry for NeuN was performed to counterstain for neurons. (c) Analogous images are shown after additional transplantation of eGFP+ neurons into the contralateral hemisphere; the Venus signal was not amplified by ICC. The arrow indicates the electroporation site; the arrowhead points out the contralateral transplantation site. The boxed area in the left hemisphere is shown in high magnification in panel 2. A GFP+ transplanted neuron (note the strong native and cytoplasmic eGFP fluorescence) is situated in the boxed area in the right hemisphere and is shown in high magnification in panels 3 and 4. While the native Venus fluorescence is well visible in the membrane of neuronal somata and proximal neuronal processes within the electroporated left hemisphere, distal axonal compartments in vicinity of the transplanted neuron within the contralateral hemisphere exhibit only weak native Venus-fluorescence. However, the tdTomato signal is well visible even in distant trans-callosal axonal compartments of electroporated neurons, tdTomato+ axon terminals (originating from contralateral recipient-derived layer II/IIICPN) are in close proximity to the transplanted neuron, suggesting that transplanted neurons might get integrated into trans-callosal long-distance circuitry within the recipient cortex. Channelrhodopsin-2-Venus (green, membrane bound), tdTomato (red, cytoplasmic), eGFP (green, cytoplasmic), NeuN (blue). Scale bars: b, c (motages), 1000 µm; b (panels 2 – 4, 50 µm; c (panel 2), 50 µm; c (panels 3,4), 100 µm.
(d) Whole-cell recordings were performed from transplanted neurons in slices of in utero-electroporated recipient brains to investigate for integration into long-distance callosal circuitry. Data for 2 example neurons are shown. ICC analysis of the slices demonstrates that the in utero-electroporations with channelrhodopsin-2-Venus and tdTomato were strictly unilateral. Arrows indicate the electroporation sites and axons of electroporated recipient-derived layer II/III CPN. Numerous channelrhodopsin-2-Venus and tdTomato positive axons are crossing the corpus callosum (CC). Arrowheads indicate the transplantation sites contralateral to the in utero-electroporation sites. Recorded neurons were filled with a biocytin-containing intracellular solution and revealed with post hoc ICC. The boxed areas of the transplantation sites are magnified and shown as pseudo-confocal stacks (collapsed into one optical plane; high magnification panels 1 – 3), displaying double-positivity of recorded transplanted neurons for eGFP and biocytin; the arrowhead within the boxed area in high magnification panel 3 indicates a recorded transplanted neuron; the blue channel (biocytin) of the boxed area is further magnified in the adjacent panel 4, displaying the fully reconstructed morphology of the recorded neuron. The complete overlap of cytoplasmic eGFP and cytoplasmic biocytin in this neuron, is shown in single optical confocal sections (panels 5, 6). Channelrhodopsin-2-Venus (green; membrane bound), tdTomato (red, cytoplasmic), eGFP (green, cytoplasmic), biocytin (blue, cytoplasmic). Scale bars: d (montages), 1000 µm; d (pseudo-confocal stacks, panels 1, 2), 40 µm; d (pseudo-confocal stack, panel 3), 70 µm; d (high magnification panel 4 and confocal panels 5, 6), 40 µm.
(e, f, top panels) Depolarization of transplanted neurons by current injections through the patch pipette resulted in robust trains of action potentials confirming neuronal identity., as expected. Evaluation of intrinsic membrane properties demonstrated the development of a mature electrophysiological phenotype with hyperpolarized resting membrane potentials (Vrest: −73 mV and −76 mV), membrane resistance- (Rm: 78 MΩ and 117 MΩ) and whole cell capacity (Cm: 166 pF and 104 pF) parameters reflecting typical values of mature pyramidal neurons. (e, middle panel) The transplanted neuron displayed in (d, left panels) was clamped to a membrane potential VC = −40mV and was continuously monitored for spontaneous synaptic events. The red and the blue boxes are resolved next to the trace on a slower time scale, revealing spontaneous EPSCs and IPSCs, indicating local and/or long-distance synaptic integration of transplanted neurons into recipient circuitry.
(e, f, bottom panels) Photo-stimulation (blue bars) of axons of in utero-electroporated recipient-derived layer II/III CPN (localized in the contralateral hemisphere) was performed while transplanted neurons were recorded in Vc = −70 mV to monitor EPSCs, or in Vc = −40 mV to monitor EPSCs and IPSCs simultaneously. (e, bottom panel) The transplanted neuron shown in (d, left panels) exhibited light-evoked IPSCs only (raw traces of 20 trials in gray; averaged traces in blue). Abolishment of the light-evoked IPSCs upon application of the AMPA/kainate and NMDA receptor blockers CNQX and APV respectively (raw traces of 20 trials in gray; averaged traces in red), strongly suggested that they were driven by glutamatergic inputs from contralateral recipient-derived layer II/III CPN to local inhibitory interneurons. The overlay of the averaged traces before (blue) and after (red) application of the blockers is shown on the right next to the traces. (f, bottom panel) The transplanted neuron shown in (d, right panels) exhibited light-evoked EPSCs (raw traces of 20 trials in gray; averaged traces in black), suggesting that the transplanted neuron received potentially monosynaptic input from contralateral layer II/III recipient-derived CPN. Also note the paired-pulse depression of light-evoked PSCs, indicating short-term plasticity of this input. (g) Epifluorescence image of the same neuron during recording. Scale bar: g, 40 µm.
Before recording from transplanted neurons, we confirmed that ChR2 was expressed at high enough levels by in utero-electroporated neurons for high-quality electrophysiology. Light flashes robustly elicited time-locked series of action potentials, and corresponding action currents (Fig. S10a), confirming appropriate expression of ChR2.
Next, transplanted neurons were patched and recorded contralateral to the in utero-electroporation site (Fig. 7g). Analysis of epifluorescence montages of entire acute slices confirmed that in utero-electroporations were performed strictly unilaterally (Fig. 7d). Preliminary recordings demonstrate that transplanted neurons developed mature electrophysiological phenotype with hyperpolarized resting membrane potentials, input resistance, and whole cell capacity parameters typical of mature cortical pyramidal neurons (Fig. 7, legend), and fired trains of action potentials upon current injection (Fig. 7e, f, upper panels).
To investigate a first level of potential functional synaptic integration, we assessed whether spontaneous excitatory and inhibitory postsynaptic currents were present in transplanted neurons. Spontaneous excitatory (sEPSCs) and inhibitory (sIPSCs) postsynaptic currents were present in all recorded neurons, reflecting spontaneous release of presynaptic excitatory and inhibitory neurotransmitters into synaptic clefts of transplanted neurons, indicating local and/or long-distance synaptic integration of transplanted neurons into recipient circuitry (Fig. 7e, middle panel).
To unambiguously determine whether transplanted neurons receive synaptic input from recipient (endogenous) neurons, in particular to investigate whether transplanted neurons functionally integrate into long-distance circuitry, we photo-stimulated axon terminals of recipient left-hemispheric CPN while recording from newly-incorporated, transplanted neurons contralaterally. In contrast to extracellular stimulation of corpus callosum, optogenetics enables analysis of trans-callosal recipient-to-transplant connectivity with laminar precision (by controlling ChR2 expression by birth-date), sub-dissecting functional connectivity established specifically from recipient (endogenous) CPN in layers II/III onto transplanted neurons in identified locations in contralateral cortex. Transplanted neurons become synaptically integrated quite frequently– 3 of 4 neurons were responsive to photo-stimulation. Importantly, based on the latencies between the onsets of the light pulses and the light-evoked postsynaptic currents, we identified excitatory connections as monosynaptic (short latencies of 4.75 ± 0.75 ms (N=2)) (Fig. 7f, bottom panel), and inhibitory connections as polysynaptic (longer latency of 6.4 ms (N=1)) which are reflecting feedforward inhibition (Fig. 7e, bottom panel). Polysynaptic transmission was investigated by application of AMPA and NMDA/kainate receptor blockers, blocking excitatory recipient CPN-to-interneuron synaptic transmission, thereby abolishing feedforward inhibition (IPSCs) of transplanted neurons (Fig. 7e, bottom panels).
Channelrhodopsin2-based mapping of efferent synaptic integration of newly incorporated neurons into complex neocortical circuitry
To ultimately achieve functional restoration of complex cortical circuitry, transplanted neurons will need to receive information from recipient brain, process inputs, and relay output to appropriate recipient CNS areas (in particular, CPN should electrophysiologically connect to recipient (endogenous) neurons in contralateral cortex).
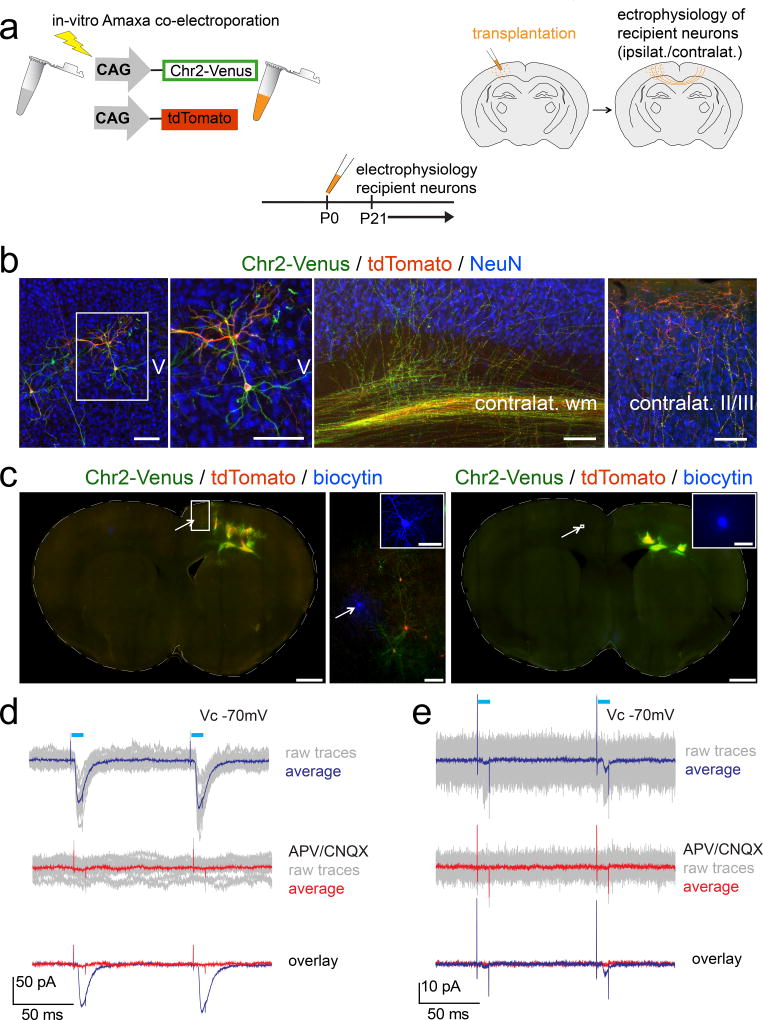
We performed transplantation of immature neurons electroporated in vitro with ChR2-Venus and tdTomato into somatosensory cortex of neonatal mice (Fig. 8a, b). A subset of transplanted immature neurons migrated away from transplantation sites, integrating morphologically into surrounding recipient cortex. Transplanted neurons extend large numbers of ChR2-Venus and tdTomato+ axons via the callosum to contralateral cortex where they innervate cortical laminae, with some axons innervating cortical layers II/III. Transplant-derived axon terminals establish close proximity to recipient NeuN+ cortical neurons, suggesting morphologically that efferent connections form between transplant-derived and recipient neurons.
Figure 8. Transplanted neurons establish functional efferent local and trans-callosal synaptic connectivity with ipsi- and contralateral recipient-derived neurons.
(a) Schematic illustrating the experimental outline: wt single neuron suspensions are in vitro Amaxa electroporated with channelrhodopsin-2-Venus (ChR2-Venus) and tdTomato constructs (left panel). Transplantation of in vitro electroporated neurons into one cortical hemisphere of wt recipients (right panel). Electrophysiology from 3 weeks of age on (bottom panel). (b, left panel) A subset of transplanted neurons migrated away from the transplantation site and entered the surrounding recipient cortex. The boxed area is shown in the adjacent high magnification panel and demonstrates channelrhodopsin-2-Venus (green; membrane bound) and tdTomato (red; cytoplasmic) co-expression by a transplanted neuron. Transplanted neurons sent large numbers of ChR2-Venus and tdTomato positive axons via the corpus callosum to the contralateral hemisphere where they invaded the cortical gray matter with a subset of axons reaching cortical layers II/III (panels 3, 4). Transplant-derived axon terminals were found in close proximity to recipient-derived NeuN+ cortical neurons suggesting functional efferent connections between transplant- and recipient-derived neurons (panel 4). ChR2-Venus (green; membrane bound), tdTomato (red, cytoplasmic), NeuN (blue). Scale bars: b, 100 µm.
(c, left montage) Post hoc epifluorescence montage and ICC analysis of an acute brain slice. The arrow marks a recorded recipient-derived neuron (biocytin filled) within the transplanted hemisphere. The boxed area is shown in the adjacent high magnification panel. Note the presence of some transplanted channelrhodopsin-2-Venus and tdTomato in vitro electroporated neurons in close proximity to the recorded recipient-derived neuron. The arrow in the high magnification panel indicates the recorded recipient-derived neuron and the inset shows a high magnification 3D confocal reconstruction (collapsed into one optical plane) of the recorded and biocytin filled recipient-derived neuron. (c, right montage) Post hoc epifluorescence montage and ICC analysis of an acute brain slice. The arrow marks a recorded recipient-derived neuron (biocytin filled) within the hemisphere contralateral to the transplanted one. Note the presence of transplanted channelrhodopsin-2-Venus and tdTomato in vitro electroporated neurons within the contralateral hemisphere. The inset shows a high magnification image of the recorded and biocytin filled recipient-derived neuron from the boxed area next to the arrow.
Electrophysiology: Recipient-derived neurons were patched and photo-stimulation (blue bars) of transplanted neurons was performed while recording at VC = −70mV. (d, e) The ipsilateral and contralateral recipient-derived neurons shown in (c, left panels) and (c, right montage), respectively, exhibited light-evoked EPSCs (d, e, top panels; raw traces of 20 and 40 trials, respectively, in gray; averaged trace in blue). Note the paired pulse facilitation, suggesting short-term plasticity. Middle panels: Abolishment of light-evoked EPSCs upon washing-in of the AMPA/kainate and NMDA receptor blockers CNQX and APV, confirmed their glutamatergic nature (raw traces in gray; averaged trace in red). Bottom panels: Overlay of the averaged traces before and after application of glutamate receptor blockers.
Note that EPSCs elicited trans-callosally (e) are about 20 times smaller in amplitude than the ones elicited intrahemispherically (d), indicating that trans-callosal connections are significantly weaker than intrahemispherical ones. Scale bars: c (montages), 1000 µm; b (high magnification), 100 µm; b (inset in high magnification panel), 50 µm; b (inset in right montage), 50 µm.
Transplanted neurons were selected by fluorescence for whole cell recording (two examples shown in Fig. S10b, c, left panels). In baseline recordings, depolarization resulted in firing of trains of action potentials, confirming appropriate neuronal maturation (Fig. S10b, c, middle panels). Photo-stimulation (Fig. S10b, c, right panels) initiated short series of action potentials (top panels), and ChR2 action currents (bottom panels) time-locked with intermittent photo-stimulation pulses, demonstrating appropriate expression levels of ChR2 by transplanted neurons. We next investigated potential ipsilateral and contralateral efferent connectivity.
Recipient neurons adjacent to transplant-derived tdTomato+ axon branches were patched and recorded ipsilateral and contralateral to transplantation sites (Fig. 8c – e). Recipient neurons were monitored electrophysiologically for synaptic input time-locked with intermittent photo-stimulation pulses (Fig. 8d, e). As expected, many contralateral recipient neurons displayed no synaptic input from transplanted neurons, providing negative controls ensuring that photo-stimulation pulses alone do not mimic synaptic responses. We recorded from 26 recipient neurons, two ipsilateral and 24 contralateral to transplantation sites. Both ipsilaterally recorded recipient neurons received synaptic input from transplanted neurons (for an example, see Fig. 8c (left montage and adjacent high magnification panel), d), while, as expected, only one of 24 contralaterally recorded recipient neurons received trans-callosal synaptic input from transplanted neurons (Fig. 8c (right montage), e). In all cases, postsynaptic currents were blocked with AMPA/kainate/NMDA receptor blockers CNQX/APV, confirming excitatory transplant-derived synapses (Fig. 8d, e). As expected, amplitudes of trans-callosal excitatory postsynaptic currents were small (average 1.9 pA), whereas amplitudes of ipsilateral excitatory postsynaptic currents averaged 50.30 ± 20.90 pA, indicating that trans-callosal transplant-to-recipient connections contribute minority input onto contralateral recipient neurons, with relatively few transplant-derived synapses projecting onto contralateral recipient-derived neurons. In contrast, ipsilateral connections are much denser, with presumably many more transplant-derived synapses projecting onto individual recipient neurons. This is consistent with results of multiple analyses using ICC and axonal fluorescence, demonstrating the development of a dense network of tdTomato+/ChR2-Venus+ axonal branches in ipsilateral transplanted cortex, with relatively fewer axons innervating contralateral cortex (data not shown).
Rare cell fusion events do not account for the observed results
Fusion of multiple immature and phagocytic cell types with recipient cells has been described– e.g. fusion of neural precursors in vitro; non-neuronal precursors with non-neuronal and neuronal precursors in vitro and in vivo; transplanted non-neuronal precursors in liver, intestine, and kidney; and monocytes/macrophages with neurons.32 Fusion is recognized as an important potential source of confusion in the absence of multiple alternative, independent assessments of cell origin as above, and has led to misinterpretation of many transplantation experiments32–34. Even though neurons identified as transplanted here display multiple identifiers confirming donor source32 (including full cellular genetic fluorescence, independent IdU/CldU labeling, progressive maturation and complexity, and appropriate temporal axon elongation), we directly investigated the theoretical possibility of cell fusion between fluorescently labeled transplanted cells/neurons (and/or phagocytosed fragments) and recipient cells/neurons by three additional, independent, complementary approaches:
First, we transplanted into Fezf2−/− mice, with complete absence of endogenous SCPN and subcerebral pyramidal tract26,30,31 (Fig. S8). Second, we performed the converse experiment, and transplanted eGFP+/Fezf2+/LacZ heterozygous or eGFP+/Fezf2LacZ/LacZ null donor neurons into wt recipients (Fig. S11). Finally, in Fezf2+/LacZ mice, b-gal serves as both reporter for Fezf2 expression and marker to distinguish transplant-derived eGFP+ wt SCPN from recipient Fezf2 heterozygous, b-gal expressing SCPN. eGFP+ wt donor neurons were transplanted into Fezf2+/LacZ recipients, and assessed by retrograde labeling from cerebral peduncle at P6 (Fig. S12).
Together, seven distinct, complementary modes of analysis rule out fusion as explaining the observed molecular and subtype fidelity, and bi-directional electrophysiological integration, of developmentally primed transplanted neurons (details in suppl. material).
Taken together, the range of molecular, connectivity, and electrophysiological data presented here indicate feasibility of reconstitution of point-to-point connectivity with precision and specificity by neurons of appropriate combinatorial molecular identity. Potentially, such point-to-point functional connectivity could include both cellular integration into correct laminar position (dendrites thus positioned relatively appropriately for correct afferent connectivity) and development of appropriate long-distance functional efferent axonal connectivity. For a graphical summary of the molecular identity data please refer to Fig. S13.
Discussion
Functional reconstruction of cortical circuitry will likely require at least partial re-establishment of appropriate and electrophysiologically functional local and long-distance synaptic connectivity. The experiments here demonstrate that transplanted immature but developmentally “primed” neurons of distinct projection neuron subtypes can integrate cellularly and positionally into postnatal cortex; maintain remarkable fidelity of differentiation and maturation– including appropriate molecular subtype identity of both subtype-specific and excluded alternate molecular controls; establish subtype-specific long-distance connectivity; and bi-directionally integrate electrophysiologically into local and long-distance circuitry.
Several groups have developed protocols for differentiating hES/mES/iPS cells into various neuron classes. These protocols, while needing further refinement, enable generation of significant quantities of neurons with features of in vivo counterparts, enabling large-scale genetic and chemical screens. These protocols also enable disease modeling with patient iPS-derived neurons, which previously relied on limited post mortem tissue35.
Recently, several studies have investigated whether ES-, iPS-, or mouse embryonic cortical-derived neurons can extend local and/or long-distance axon projections after transplantation into early postnatal or lesioned adult mouse brains18,20,21,22,23,36,37. These studies mainly relied on transplantation of large numbers of neurons or progenitors, resulting in formation of isolated heterotopias, spanning recipient cortical gray and white matter, with only rare cells exiting heterotopias and entering surrounding cortex. While these studies report that trans-callosal and corticofugal projections can be established by transplants, retrograde labeling in some studies reveals that projections originate essentially only from neurons within heterotopias, not from the rare transplanted neurons that disperse into surrounding recipient cortex21,22. Thus, neuronal dendrites and afferent connections are largely isolated within heterotopias, reducing likelihood that neurons become appropriately integrated as individual cells. Further, projections in these studies often appear quite multidirectional exiting heterotopias, raising the possibility that projections passively follow and fasciculate with axon tracts where cells are placed, in direct contact38. In studies analyzing some transplanted neurons by ICC for subtype marker expression22, e.g. CTIP2 for subcerebral projection neurons, analyses remained restricted to neurons within isolated heterotopias, limited to a single marker at a time, rather than combinations of alternative, exclusionary, and progressively refined markers simultaneously on individual neurons. These limitations leave it unclear whether observed results might be due to 1) potentially “stalled”, immature, and/or abnormal, “hybrid”, and/or unrefined differentiation in case of hES/mES/iPS-derived neurons39,40; 2) potential inadequacy of permissive and/or instructive signals within recipient cortex to enable subtype-specific differentiation with specific circuit formation; and/or 3) potential failure of newly incorporated neurons to effectively respond to axon guidance cues and/or subtype-specific fasciculation signals that might enable appropriate circuit formation.
To investigate whether cellular level integration of dispersed single neurons of defined subtype identity, with preferential positioning in appropriate cortical layers, and with establishment of appropriate efferent axon projections can be achieved (including to long-distance trans-callosal and subcerebral targets), we performed micro-transplantation of immature cortical neurons followed by ICC for alternative sets of 2 or 3 transcriptional controls, combined with retrograde neuroanatomical labeling of axon projections.
Further experiments identified optimal stages of origin of transplanted cells to generate integrated neuron subtypes, and to investigate whether cells are more likely to acquire 'correct' fate by maintaining a prior committed fate rather than by switching fates. Experiments involving double birthdating by IdU and CldU within donor embryos reveal that donor neurons successfully integrating and projecting to subcerebral targets became postmitotic >16 hours prior to transplantation, so they were neither progenitors nor immediately “newborn” neurons when they migrated and integrated. Analysis of stage-matched littermate cortex in vivo identified that corresponding postmitotic neurons already formed deep cortical layers, already CTIP2+. These data indicate that early postmitotic DCX+ neuroblasts destined to form deep-layer projection neurons, after assuming appropriate positions in cortical plate when dissociated, account for most neurons integrating into recipient cortex, and establishing subcerebral projections after transplantation. Thus, a “fate-switch” of presumptive upper layer CPN toward SCPN identity seems not to occur, since upper layer CPN are not yet born at this stage from SVZ progenitors. Together, these experiments narrow and substantially define optimally integrating cells to early postmitotic neurons, whether largely fate-restricted deep layer projection neurons, or still plastic postmitotic neuroblasts with partial fate-restriction.
These conclusions are further supported by additional data (Fig. S11, demonstrating that genetic deletion of SCPN from the donor cells using Fezf2−/− mice results in absence of transplant-derived subcerebral projections. With Fezf2−/− donors, neither CPN nor other projection neuron populations form (alternate, switched) subcerebral projections, neither “confused” nor with mixed identity.
Effective regeneration and repair of circuitry likely depends on the extent of restoration of synaptic, electrophysiological connectivity. To investigate bi-directional functional synaptic connectivity established between transplant-derived neurons and recipient layer II/III CPN in ipsilateral or contralateral cortex, we performed optogenetic-based electrophysiology in acute brain slices of recipients. In some experiments, optogenetic-based electrophysiology selectively stimulated projections of transplant-derived neurons. This enabled direct investigation of potential local and long-distance functional output connectivity established by transplanted neurons with recipient neurons, not only local connectivity (standardly accessible via paired recordings in prior studies). Our data identify that appropriate, even inter-hemispheric functional restoration of cortical circuitry can be achieved.
Understanding molecular mechanisms underlying cellular migration, dendritic and axonal growth and guidance, target selection, and synaptogenesis might aid efficient integration of new neurons into more mature recipient brains. Our data indicate that optimally successful integration depends on transplantation of relatively small numbers of neurons, enabling them to overcome reciprocal attraction (shown by Ladewig et al.41 to be involved in limiting migration of hESC-derived neurons) and migrate into recipient cortex. Micro-transplantation of small numbers of cells naturally results in few SCPN or CPN per brain. In vitro directed differentiation and/or enrichment for postmitotic projection neuron subpopulations could potentially increase directed connectivity. While CNS development is ongoing early postnatally, and instructive and permissive signaling (including but not limited to axon guidance molecules, guidepost cells, and dendritic arborization signals) is likely to be declining but still active, it is clear from prior studies that long-distance connectivity is possible even in adult CNS. Axons of transplanted neurons can be directed in adult recipient brain by virus-mediated re-expression of neuron-specific chemoattractant and chemorepellant molecules42. Other data demonstrate establishment of projections to distant targets after embryonic mouse neuron micro-transplanation12,13,14, or following transplantation of mESC-derived neurons22 or blocks of embryonic tissue43.
Our experiments indicate that neocortical connectivity can be reconstituted with point-to-point precision, including cellular integration of correct molecular subtypes into correct laminar positions, development of appropriate long-distance projections, and functional afferent and efferent local and long-distance connectivity. Data suggest that micro-transplantation of primed cortical neurons within mature brain might similarly enable reconstruction of complex neocortical circuitry. It will be of interest to further investigate circuit specificity of such connectivity, and whether axon-pathfinding is directed by re-expressed guidance cues and fasciculation signals, and/or by pre-existing and degenerated axon tracts with potentially residual myelin sheaths acting as passive guidance structures for transplant-derived axons. Results have implications both toward repair of complex circuitry and regarding directed differentiation of progenitors/stem cells for in vitro mechanistic modeling and therapeutic screening maintaining precision of relevant neuron subtypes.
Methods
Animals
All experimental mouse studies were approved by the Harvard University IACUC, and were performed in accordance with institutional and federal guidelines. Wild-type C57BL/6 mice were purchased from Charles River Laboratories (Wilmington, MA); eGFP-expressing transgenic mice (C57BL/6-Tg(ACTB-EGFP)1Osb/J)44 were purchased from the Jackson Laboratories (Bar Harbor, ME). This transgenic mouse line expresses an "enhanced" GFP (eGFP) driven by the chicken beta-actin promoter and cytomegalovirus enhancer, in all tissues except erythrocytes and hair44. Fezf2 null (Fezf2−/−) mice, Fezf2LacZ/LacZ and Fezf2AP/AP, mice were generated by Hirata and colleagues45 and Chen and colleagues31, respectively. For timed gestations, the morning of initial vaginal plug observation is defined as embryonic day E0.5. The day of birth is designated postnatal day 0 (P0). The genotypes of Fezf2 mutant mice were identified by PCR on tail/brain tissue genomic DNA, as described previously30,31,45. Genotypes of eGFP transgenic mice were determined using brief fluorescence excitation on live pups.
All experiments involving in utero-electroporation and/or transplantation of Amaxa electroporated neurons were performed in wild-type (wt) CD1 background mice; these timed-pregnant mice were purchased from Charles River Laboratories (Wilmington, MA).
Male and female mice were used as donors and recipients. A standard 12h light / 12h dark cycle was maintained in animal housing facilities.
Micro-transplantation to produce physical chimeras, and retrograde labeling
We performed these experiments in young postnatal mice to enable the most subtype-specific molecular analyses. Almost all of the most informative molecular controls/markers become increasingly non-specific starting a few days after birth, posing significant limitations on such studies in mature CNS. Therefore, we performed experiments in young mice so transplantation, neuronal migration and maturation, axonal projection, and analyses by combined retrograde labeling and ICC could be completed during this period of marker subtype-specificity.
Transplantation was performed at P0/P1, and injections of retrograde axonal labels were performed 5–6 days post-transplantation (the earliest time following axon extension to the cerebral peduncle or the contralateral hemisphere – as documented in Figure S1). ICC was performed 2 days after retrograde label injection; both CTB555 and LumaFluor microspheres are retrogradely transported to the soma within this time.
We crossed male eGFP transgenic mice with either wt female C57BL/6 mice or Fezf2+/LacZ mice, and obtained embryonic day 13.5–15.5 (E13.5–E15.5) eGFP-labeled embryos as donor neuron sources (spanning the peak days of birth of subcerebral projection neurons (SCPN) and callosal projection neurons (CPN)). On the day of transplantation, eGFP+ E13.5–15.5 embryos were selected by fluorescence excitation, brains were dissected out, and cortices were dissociated to obtain single cell suspensions. Cortices were dissected in ice-cold dissociation medium (20 mM glucose, 0.8 mM kynurenic acid, 0.05 mM dl-2-amino-5-phosphonopentanoic acid (APV), 50 U/ml penicillin-0.05 mg/ml streptomycin, 0.09 M Na2SO4, 0.03 M K2SO4, and 0.014 M MgCl2; pH 7.35±0.02), and enzymatically digested in dissociation medium containing 0.16 mg/l l-cysteine HCl, 10 U/ml papain (Worthington, Lakewood, NJ), and 20 U/ml DNAse (pH 7.35 ± 0.02) at 37 °C for 15–20 min, followed by rinsing with dissociation medium containing OVO/BSA (pH 7.35±0.02) at RT to inhibit the papain, and finally washed with ice-cold OptiMEM (GIBCO, Life Technologies, Gaithersburg, MD) containing 20 mM glucose, 2.5% fetal bovine serum (Invitrogen, Carlsbad, CA), and both 0.4 mM kynurenic acid and 0.025 mM APV to protect against glutamate-induced neurotoxicity46. Cortices were mechanically dissociated by gentle trituration, and single cortical cells were re-suspended at about 7–15 thousand living (trypan blue negative) cells/µl. Using established methods1,13,14, P0–P1 wt, Fezf2AP/AP; Fezf2LacZ/LacZ; or Fezf2+/AP; Fezf2+/LacZ mice were anesthetized by hypothermia, and dissociated cortical cells were micro-transplanted into sensorimotor cortex at 9 sites with a digitally controlled nanoinjector (Nanoject II; Drummond Scientific, Broomall, PA) using pulled glass micropipettes with a tip diameter of ~50–60 µm. Dissociated cells were micro-injected at these 9 sites (~300–700 cells/site; 4.6 nl/deposit, 10 deposits/site) in “columns”, from 450–500 µm depth to the pial surface in 50 µm steps. Typically, only a few percent of the cells injected actually remain within the parenchyma. Most are extruded via tissue compressive pressure from each of these relatively elastic microcavities. This approach enables some cells, presumably those on the periphery, to enter the parenchyma and undergo integration. We find, following transplantation of ~3000–7000 total cells injected, only a few hundred total cells remain across the 9 sites. Following monitored recovery, pups were returned to their dams. At P6 (5–6 days post-transplantation), the transplanted (physical chimera) mice were retrogradely labeled from either the contralateral hemisphere or the cerebral peduncle with red fluorescent microspheres (LumaFluor) or Alexa555-conjugated cholera toxin subunit B (CTB555) (contralateral: stereotaxic surgery using a Cunningham neonatal mouse adaptor fit to a standard stereotaxic frame (Stoelting); cerebral peduncle: ultrasound-guided, using a Vevo 770 ultrasound backscatter microscopy system (VisualSonics)). Pups were perfused at P8. Tissue was processed as described in the histology Methods section below.
Optimization of micro-transplantation
We largely avoided heterotopia formation by micro-transplantation of limiting, small cell numbers. Toward this goal, pilot experiments were carried out to determine the most appropriate recipient age, cell concentration, and transplantation depth, enabling transplantation and dispersed neuronal integration into cortical gray matter with only minimal heterotopia formation.
These pilot experiments confirmed prior findings that high-concentration transplants result in massive heterotopia formation, extending from cortical gray to white matter, and, in some cases, even into more basal structures (data not shown). Many neurons even within heterotopias generally have the ability to project to long-distance targets (data not shown); however neurons transplanted into myelinated tracts have previously been demonstrated to grow long interfascicular axons promiscuously, without specificity.38 In subsequent experiments, the transplantation procedure was optimized stepwise by (i) lowering the cell concentration of the transplantation mix, and (ii) adjusting the transplantation depth, aiming at avoiding placement of cells into the white matter, or into more ventral structures (e.g. striatum). This iteratively optimized procedure resulted in a marked reduction of the size of cell aggregations. Finally, micro-transplantation of even smaller cell numbers enabled integration of transplanted neurons as single cells into recipient cortical gray matter (and largely abolished heterotopia formation within cortical gray matter), with integration frequently even far away from transplantation tracks. Concentrations around 10 thousand cells/µl and lower were found to optimally enable single cell/neuron integration. Using these conditions, a large proportion of micro-transplanted neurons dispersed into surrounding recipient cortex, with migration distance ranging from several tens up to several hundred micrometers away from transplantation tracks. Newly incorporated neurons integrated throughout cortical layers, with the majority assuming positions in or ventral to layer V, and developed complex and remarkably normal pyramidal morphology and dendritic growth. Both neurons close to transplantation tracks (including neurons within heterotopias in initial pilot experiments) and neurons that migrated far away from other transplanted neurons were identified with projections to long-distance targets. Therefore, we designed all subsequent experiments to minimize and optimally entirely avoid heterotopia formation.
Prior reports have demonstrated that transplanted neurons (even of non-cortical origin) misplaced in cortical white matter extend axonal projections (even in adult recipients), passively following pre-existing fiber tracts38. Therefore, to ensure that transplanted neurons had full, autonomous choice as to where to project their axons (unbiased by pre-existing recipient axon tracts e.g.) molecular analysis combined with retrograde labeling was a priori focused on eGFP+ transplanted neurons integrated into cortical gray matter, and separated from cortical white matter by at least one layer of recipient neurons.
Micro-transplantation of very small quantities naturally results in transplantation of only very few SCPN or CPN per brain. As noted above, following transplantation, only a few hundred total transplanted neurons per brain remain across the transplantation sites. The cellular composition can be at least partially estimated from prior work and the current experiments. At E15.5, ~6% of all cells in the donor cell suspension can be expected to be SCPN, and ~5% CPN (according to FACS purification data of dissociated E15.5 somatosensory cortex by Molyneaux et al.47).
This sparse neuronal integration enabled single neuron resolution analysis of projecting axons, identified by retrograde labeling. Since retrograde labeling depends on the target injection size and distribution volume, the typically ~3 dispersed transplanted neurons of each distinct subtype within cortical gray matter identified by retrograde labeling to project to long-distance targets is well within expectations.
Double birthdating of donor neurons prior to transplantation and retrograde labeling
Transplanted eGFP+ neurons were first double birthdated in utero, then retrogradely labeled from the cerebral peduncle. All cortical neurons born from E11.5 onward were pre-labeled with IdU by continuous administration in the drinking water (1mg/ml) to pregnant dams. From E14.5 onward (~16 hours before transplantation), all newly born neurons were additionally labeled with CldU (17.1mg/ml) by 2 subsequent CldU i.p. injections (2.5µl/g body weight of the pregnant dam; interval of 2 hours) and continuous administration of IdU and CldU in the drinking water (IdU 1mg/ml; CldU 1mg/ml) to enable two-tiered temporal analysis.48
Some transplanted (but not retrogradely labeled) neurons within analyzed sections were found to be CldU+, thus serving as internal positive controls for CldU immunocytochemistry.
Expression of channelrhodopsin2-Venus (ChR2-Venus) in recipient mice via in utero electroporation
A vector containing channelrhodopsin2-Venus (pACAGW-ChR2-Venus-AAV) under the control of a constitutively active CMV/beta-actin promoter was used49 (purchased from Addgene, ID 20071). A vector containing IRES-eGFP under the control of a constitutively active CMV/beta-actin promoter (CBIG) (generous gift of C. Lois, Caltech, Pasadena, CA) was modified, and the eGFP open reading frame was replaced by the coding sequence of the fluorescent protein tdTomato (CBIT).
We mixed ChR2-Venus (final conc. 1.5 µg/µl) and tdTomato constructs (final conc. 1 µg/µl) with 0.005% Fast Green (for visualization), injected the mixture in utero into a lateral ventricle of CD1 embryos at E14.5, and electroporated cells in the dorsal ventricular zone, as described previously30,50. Only some pups were manipulated, while their littermates were left unperturbed to optimize survival and in utero injection success.
Expression of channelrhodopsin2-Venus (ChR2-Venus) and either tdTomato or eGFP in donor neurons by Amaxa electroporation
ChR2-Venus construct (7–8 µg; see section above) and 7–8 µg of either CBIT or CBIG construct Maxi Prep DNA was mixed with 100 µl nucleofector solution for Amaxa electroporation of mouse neurons (Lonza).
Single cell suspensions were centrifuged at 1000 RPM at 4°C for 7 minutes, then re-suspended in the nucleofector-DNA mix. Amaxa electroporation was performed in cuvettes provided with the mouse neuron Amaxa electroporation kit (Lonza), using program O-005. At the end of the electroporation, 900 µl of ice-cold OptiMEM (GIBCO, Life Technologies, Gaithersburg, MD) containing 20 mM glucose, 2.5% fetal bovine serum (Invitrogen, Carlsbad, CA), and both 0.4 mM kynurenic acid and 0.025 mM APV was immediately added to minimize the exposure time of dissociated neurons to the nucleofector solution. Cells were centrifuged again, resuspended in a final volume of ~50 µl, and transplanted within ~120–150 minutes. Cell suspensions were kept on ice until all transplantation was complete.
Transplantation to produce physical chimeras; optogenetics in chimeras
Afferent integration of transplanted neurons
E14.5 CD1 wt embryos were electroporated in utero with constructs encoding ChR2-Venus and tdTomato (see sections above).
On P0–P1, pups were screened for cortical electroporation sites (since not all pups in each uterine horn were electroporated, due to positioning, and to optimize litter care; tdTomato fluorescence enabled screening without exciting ChR2). Only pups with strictly unilateral cortical electroporation sites were selected. Recipient mouse pups were anesthetized by hypothermia. The skin was opened with micro-scissors, and moistly maintained as a flap. The exposed skull was illuminated under a fluorescence dissecting microscope, and the area of tdTomato fluorescence was marked on the skull to guide subsequent transplantation.
Transplantation of E17.5 wt CD1 cortical cell suspensions, following eGFP Amaxa electroporation ex vivo, was performed into the contralateral non-electroporated cortex, covering the area homotopic to the in utero-electroporation site. E17.5 was selected as the donor age to bias for postmitotic layer II/III CPN, and to minimize the formation of donor-derived heterotopic cell aggregates within the recipient cortex (as is typically observed with large numbers of mitotic progenitors in preparations from early embryonic ages).
Efferent integration of transplanted neurons
E14.5 CD1 wt cortical suspensions were Amaxa electroporated with ChR2-Venus− and tdTomato-encoding constructs. Neurons were transplanted into the right cortical hemisphere of P0–P1 recipient CD1 wt pups.
Electrophysiology
At 4–6 weeks of age, transplanted recipient mice were deeply anesthetized and decapitated. Brains were quickly dissected out of the skull. 300 µm thick acute slices were prepared with a Leica VT 1000 S vibrating microtome. Brains were sectioned in ice-cold artificial CSF (aCSF) containing 83 mM NaCl, 2.5 mM KCl, 3.3 mM MgSO4, 1 mM NaH2PO4, 26.2 mM NaHCO3, 22 mM glucose, 72 mM sucrose, and 0.5 mM CaCl2, equilibrated with 95% O2/5% CO2. Slices were allowed to equilibrate for 20 min at 37°C, followed by 1h at room temperature before placement into a recording chamber, where they were continuously perfused at a constant flow rate of 2ml/min with normal aCSF containing 119 mM NaCl, 2.5 mM KCl, 1.3 mM MgSO4, 1 mM NaH2PO4, 26.2 mM NaHCO3, 22 mM glucose, and 2.5 mM CaCl2, equilibrated with 95% O2/5% CO2. Micropipette resistance ranged between 5 and 7 MOhm. Series resistance was not compensated.
Electrophysiological recordings were performed at room temperature within 5 hours of slice preparation. For voltage clamp recordings, we used Cs-gluconate-based internal solution containing 130 mM D-gluconic acid, 130 mM CsOH, 5 mM NaCl, 10 mM HEPES, 12 mM phosphocreatine, 3 mM MgATP, 0.2 mM NaGTP, 1 mM EGTA, and 5 mg/ml biocytin51. For current clamp recordings, we used a K-gluconate-based internal solution containing 130 mM KCl, 10 mM HEPES, 10 mM phosphocreatine, 3 mM MgATP, 0.5 mM NaGTP, 0.2 mM EGTA, 2.5 mM glutamate, and 5 mg/ml biocytin. AMPA receptor-mediated EPSCs were recorded by holding cells at −70 mV, whereas mixed AMPA and NMDA receptor-mediated EPSCs were recorded at +40 mV. GABAA receptor-mediated IPSCs were recorded at 0mV, and for some cells both EPSCs and IPSCs were recorded at −40mV.
ChR2 was activated in distant contralateral axons and synapses far from their neuronal somas, within the entire optical field of view, using a custom-built LED-based illuminator.52 A high-intensity LED array (5–10 mW/mm2; CBT-120B, Luminus Devices) was coupled to the rear lamp-housing of an Olympus BX51 upright microscope. Occasional light stimulation transients (from the LED power source) were easily distinguished from synaptic currents, and were not affected by synaptic blockers.
Neurotransmitter-specific blockers of synaptic transmission were washed in with a custom-made perfusion system, and were used at the following concentrations: 10µM CNQX; 100µM APV. Addition of biocytin to the intracellular solution enabled post hoc morphological analysis of recorded transplant- or recipient-derived neurons. Slices were fixed in 4% paraformaldehyde (PFA) at 4°C overnight, then rinsed in phosphate-buffered saline (PBS). Immunocytochemistry on thick sections was performed as described below, except that incubation with primary antibodies was performed at room temperature. Post hoc analysis was performed for each acute slice to confirm that cortical in utero-electroporation sites were strictly unilateral.
Histology and immunocytochemistry
Following deep anesthesia with Avertin, mice were transcardially perfused with 0.1 M PBS, pH 7.4, followed by 4% PFA in PBS buffer, and brains were dissected out of the skulls. Brains were post-fixed in 4% PFA overnight at 4°C, followed by 3 PBS rinses, then coronally or sagittally sectioned at 50 µm thickness with a vibrating microtome (Leica). For sagittal sections, brains were embedded in 4% low melting temperature agar (Sigma-Aldrich) before sectioning. Vibrating microtome sections were collected into PBS or PBS containing 0.05% sodium azide (Sigma-Aldrich), incubated in blocking solution (PBS, 0.05% sodium azide, 0.3% BSA, 8% goat serum, and 0.3% TritonX-100) for 30 min at room temperature, then incubated in primary antibody: rabbit anti-GFP IgG (1:500, Invitrogen, A-11122); chicken anti-GFP IgG/IgY (1:500, Abcam, ab13970); rat anti-CTIP2 IgG (1:250, Abcam, ab18465); mouse anti-SATB2 IgG (1:200, Abcam, ab51502); rabbit anti-FOG2 IgG (1:250, SantaCruz Biotech., sc-10755); rabbit anti-b-gal IgG (1:1000, MP Biomedical, 0855976); rabbit anti-DCX IgG (1:1000, Sigma, D9943); mouse anti-NeuN IgG (1:200, Chemicon, MAB377); mouse anti-BrdU IgG (1:500, BD, 347580); rat anti-BrdU IgG (1:500, Accurate Chemical, OBT0030); rat anti-L1 IgG (1:500, Chemicon/Millipore, MAB5272); mouse anti-synaptophysin IgG (1:250, Chemicon, MAB5258) overnight at 4°C. Sections were washed three times in PBS for 10 min, then incubated in secondary antibody: Alexa 488 goat anti-rabbit IgG (1:500, Invitrogen, A-11034); Alexa 488 goat anti-chicken IgG antibody (1:500, Invitrogen, A-11039); Alexa 405 goat anti-mouse IgG (1:500, Invitrogen, A-31553); Alexa 647 goat anti-rat IgG (1:500, Invitrogen, A-21247); Alexa 647 goat anti-rabbit IgG (1:500, Invitrogen, A-21244); Alexa 546 goat anti-rat IgG (1:500, Invitrogen, A-11081); Alexa 594 goat anti-rat IgG (1:500, Invitrogen, A-11007) for 2 h at room temperature. Sections were mounted on glass slides after washing 3 times for 10 minutes with PBS. Immunocytochemistry (ICC) for BrdU was preceded by 2 h treatment with 2N HCl at room temperature for antigen retrieval. All primary and secondary antibodies used in this paper have been extensively used in prior studies by our and other laboratories (e.g. references 26 – 30).
Since CTIP2, SATB2, and FOG2 become increasingly non-specific with time (e.g. CTIP2 starts to be expressed by interneurons from 1 week of age, and is also increasingly expressed in upper layers) we performed ICC at the latest optimal time for cellular identification without loss of specificity (P8).
Image acquisition
Images were captured on a fluorescence microscope (Nikon E90i or Nikon E1000) equipped with a Nikon Intensilight C-HGFI source or an X-Cite 120 illuminator (EXFO), respectively, using high numerical aperture optics and a cooled CCD camera (ANDOR or Q-Imaging, respectively) running NIS Elements software (Nikon) or Velocity software (Improvision, Lexington, MA).
Five-channel confocal imaging was performed at the Harvard Center for Biological Imaging on a Zeiss 710 inverted confocal microscope equipped for spectral unmixing and for acquisition of confocal montages of entire tissue sections, running ZEN software (Zeiss). Other confocal images were obtained using a BioRad Radiance 2100 Rainbow laser-scanning confocal system based on a Nikon E800 microscope with LaserSharp 2000 imaging software (Bio-Rad Laboratories). Some images were subsequently processed for 3D reconstruction in Imaris (Bitplane). For an optimal visual reproduction of the data, images were adjusted for contrast, brightness, color balance, and size in Photoshop CS3 (Adobe, San Jose, CA), and assembled in Power Point for Mac 2011 (Microsoft Corporation, Redmond, WA), and in Illustrator CS3 (Adobe, San Jose, CA).
Statistics and Reproducibility
Retrograde labeling experiments
For analysis of molecular identity (CTIP2, SATB2, FOG2 ICC) and for double birthdating experiments combined with retrograde labeling (from cerebral peduncle or contralateral hemisphere) all neurons found throughout all experiments and complying with the a priori criteria (neurons located in cortical gray matter separated from white matter by at least one layer of recipient derived cortical neurons and retrogradely labeled from target sites) were included in analysis (therefore no randomization was performed); no neurons meeting these criteria were excluded. Data collection and analysis were not performed blind to the conditions of the experiments.
Fig. S13 provides a summary graphical representation of the quantification of the molecular identity data that integrates the individual quantifications in all figures depicting combined retrograde labeling and molecular identity data. Further, we performed statistical analysis using Fisher’s exact test (please also refer to the legend to Fig. S13) demonstrating a remarkable level of molecular-to-projection fidelity, with a p-value <0.00001 (exact p-value: 0.0000020104, regardless whether calculated with one or two tails). Fisher’s exact test is explicitly valid when a chi-squared test-based approximation is inadequate, since sample sizes are small, or the data are very unequally distributed among the cells of a table.
Experiments involving transplantation and retrograde labeling (excluding pilot experiments to optimize transplantation depth and cell numbers) employed 15 litters with on average 6–8 pups (in total ~100 pups). Each pup underwent a multi-step experimental sequence with transplantation followed by retrograde labeling. Retrogradely labeled neurons presented derive from 27 successfully injected pups. In accordance with standard procedures in the field for such data, more than 3 neurons per condition were analyzed for core data; no further formalized statistical methods were used to predetermine sample sizes. In general, multiple aspects of neuronal identity are interrogated via multiple molecular, birthdate, and/or connectivity analyses, so overall numbers are more in the dozen+ range for integrated interpretation of results for any neuronal subtype identity and its fidelity of connectivity. These sample sizes, though small, provided power for exceptionally significant and robust statistical analysis.
Additional mice were used for electrophysiology and other experiments not involving retrograde labeling.
Experimental Numbers
A. Retrograde labeling experiments
I: Progressive maturation of transplanted neurons visualized by retrograde labeling; long-term maintenance of axon projections (as long as assessed; >5 weeks)
ICC for DCX and NeuN of 5 eGFP+ retrogradely labeled neurons is presented in Fig. 2, Fig. S2, and Fig. S3: 2 neurons at 1 week, 2 neurons at 2 weeks, and 1 neuron at 5 weeks. We analyzed additional neurons at 1 week, 3 weeks, and 5 weeks, and confirmed the same findings. Fig. 2, Fig. S2, and Fig. S3 show progressive maturation of transplanted and retrogradely labeled SCPN and CPN over a time course of ~5 weeks according to DCX and NeuN immunolabeling. Retrograde labeling displayed in Fig. S3, was performed at P35 (CTB555 injection was performed 5 weeks post-transplantation, rather than 5–6 days post-transplantation, as in all other experiments). These data indicate that axons and axon integrity remained intact at least up to this time post-transplantation.
Total number of neurons analyzed: 1 week analysis, n=4; 2-week analysis, n=2; 3-week analysis, n=3; 5-week analysis, n=4. ICC for DCX and NeuN of 5 eGFP+ retrogradely labeled neurons (from cp or contralateral hemisphere with CTB555) is displayed from 2 neurons at 1 week, 2 neurons at 2 weeks, and 1 neuron at 5 weeks:
Fig. 2b: 1 SCPN, analysis for DCX and NeuN at 8 days post-transplantation.
Fig. S2b: 1 CPN, analysis for DCX and NeuN at 7 days post-transplantation.
Fig. 2c: 1 SCPN, analysis for DCX and NeuN at 14 days post-transplantation.
Fig. S2c: 1 CPN, analysis for DCX and NeuN at 14 days post-transplantation.
Fig. S3: 1 CPN, analysis for DCX and NeuN at P37/P38 (~5 weeks post-transplantation); retrograde labeling was performed at P35 rather than 5–6 days post-transplantation.
2 additional neurons at 1 week, 3 additional neurons at 3 weeks, and 3 additional neurons at 5 weeks were analyzed, confirming the same findings.
II: Double birthdating of transplanted neurons, combined with retrograde labeling
Total n=4 neurons analyzed with combination double IdU/CldU birthdating plus retrograde labeling. Fig. 3 depicts double birthdating data for 1 transplanted and retrogradely labeled SCPN (IdU+/CldU−). We analyzed 3 additional neurons; all were IdU+/CldU− (Fig. S4).
III: Analysis of molecular identity of transplanted neurons combined with retrograde labeling
These are presented in Figures 4, 5, 6, and Figures S5, 6, 7, 8, 12.
Representative neurons are depicted in the main figures, and all additional analyzed neurons in the Supplementary Figures. As noted above, all neurons meeting the a priori criteria were included in the analysis; none were excluded.
IIIa: CSMN/SCPN (analysis of n=13 transplanted neurons)
Fig. 4b and Fig. S5a: analysis for CTIP2 and SATB2; n=4.
Fig. 4c and Fig. S5b: analysis for FOG2 and Dapi; n=4.
Fig. 4d: five-channel ICC, analysis for CTIP2, FOG2, and SATB2; n=1.
Fig. 6: five-channel ICC, analysis for CTIP2, SATB2, and Fezf2; integration into appropriate cortical layer V; n=1.
Fig. S6: analysis for high- vs. low-level CTIP2 expression by epifluorescence microscopy in addition to confocal microscopy (as performed in Figs. 4, 6, and Fig. S5); n=1.
Fig. S12: cell fusion experiment; Fezf2+/b-gal recipient: five-channel ICC, analysis for CTIP2+, SATB2−, and b-gal− transplanted eGFP+ neuron retrogradely labeled from the cp; n=1.
Fig. S8: Transplantation into Fezf2 null recipient: analysis for CTIP2 and SATB2; n=1.
IIIb: CPN (analysis of n=9 transplanted neurons)
Fig. 5b and Fig. S7a: analysis for SATB2 and CTIP2; n=4.
Fig. 5c and Fig. S7b: analysis for SATB2 and FOG2; n=4.
Fig. 5d: five-channel ICC, analysis for SATB2, CTIP2, and FOG2; n=1.
B. Experiments not involving retrograde labeling
Total numbers: 3 SCPN; 2 CThPN; 2 CPN. 1 SCPN, 1 CThPN, and 2 CPN are presented in Fig. 1.
Fig. S1 illustrates progressive axon extension. Axons of transplanted neurons reach the cerebral peduncle or the contralateral cortical hemisphere by ~5 days post-transplantation, enabling all retrograde labeling experiments at P5/P6. These experiments also confirm that axons reach the cerebral peduncle or the contralateral hemisphere by 5–6 days post-transplantation.
Fig. S9 shows 1 transplanted neuron surrounded by synaptophysin staining.
Fig. S11 shows data obtained at 3 weeks post-transplantation for subcerebral projections (and 2 weeks post-transplantation for callosal projections; see inset). Neurons analyzed earlier, at 1 and 2 weeks post-transplantation, confirm the 3 week findings presented in Fig. S11; these were omitted from the figure due to space constraints.
Electrophysiology
Fig. 7e and f, top panels, n=4 (2 examples shown out of 4 total).
Fig. 7e, middle panel, n=6 (1 example shown out of 6 total).
Fig. 7e and f, bottom panels, n=4 (3 out of 4 recorded neurons were responsive to photo-stimulation: e, bottom panel, n=1 (shown); f, bottom panel, n=2 (1 of 2 examples shown)).
Fig. 8 26 recipient-derived neurons were recorded, 2 ipsilateral and 24 contralateral to the transplantation sites. 2 out of 2 ipsilaterally recorded neurons received synaptic input from transplanted neurons (one example shown in c (left montage and adjacent high magnification panel), d); 1 of 24 contralaterally recorded recipient neurons was found to receive trans-callosal synaptic input from transplanted neurons (shown in c (right montage), e).
Fig. S10: Total n=7:
Fig. S10a, n=1 (1 example shown).
Fig. S10b, c, n=6 (2 examples shown).
Life Sciences Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary.
Data availability
All core data generated or analyzed during this study are included in this published article (and its supplementary information files). The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary Material
Figure S1: Time course of subcerebral and trans-callosal axon extension by transplanted neurons. Transplanted neurons progressively extend long-distance axon projections and reach pons and more distal subcerebral targets as well as the contralateral cortex (f and g) by 5–6 days post-transplantation (dpt). Subcerebral axon projections: 4 dpt (a [striatum] and b [cerebral peduncle]), 5 dpt (c [pons] and d [proximal medulla oblongata]), 6 dpt (e [distal medulla oblongata]). Trans-callosal axon projections: 4 dpt (h), 5 dpt (i), 6 dpt (j). eGFP (green); L1 (red). Scale bars: a – e, 200 µm; h, – j, 100 µm.
Figure S2 (related to Fig. 2): Transplanted neurons assuming positions in cortical layers II–VI progressively mature and successfully extend trans-callosal long-distance axon projections. (a) Schematic illustrating the experimental outline. Micro-transplantation of eGFP+ single cell suspension into P0/P1 recipient sensorimotor cortex. Stereotaxic injection of retrograde axonal tracer (CTB555) into the contralateral hemisphere at P6. Analysis at P7 and P14. (b, c) A subset of transplanted neurons project trans-callosally. These neurons progressively mature from a DCX+ and NeuN−/low (b) to a DCX−/low and NeuN+ state (c). Compare the DCX expression of the recipient cortex at both time points. DCX decreases dramatically on a very similar time scale in the recipient cortex as for the transplanted neurons. The positions of the boxes in low magnification panels (b, c) correspond to the cortical area examined in high magnification panels. Arrows indicate transplanted neurons. eGFP (green), CTB555 (red), DCX (immature neuronal marker; purple), NeuN (mature neuronal marker; blue). Scale bars: b, c (low magnification), 50 µm; b, c (high magnification), 10 µm.
Figure S3 (related to Fig. 2): Long-distance axon projections of transplanted neurons remain stable long-term. (a) Schematic illustrating the experimental outline. Micro-transplantation of eGFP+ single cell suspension into P0/P1 recipient sensorimotor cortex. Stereotaxic injection of retrograde axonal tracer (CTB555) into the contralateral hemisphere at P35 and analysis at P37/P38. (b) A subset of transplanted neurons establish trans-callosal axon projections, remaining stable long-term, without pruning or retraction for periods as long as analyzed (5 weeks). These neurons progressively mature to a DCX− and NeuN+ state. The position of the box in the low magnification panel (b) corresponds to the cortical area examined in high magnification panels. Arrows indicate a transplanted neuron. eGFP (green), CTB555 (red), DCX (immature neuronal marker; purple), NeuN (mature neuronal marker; blue). Scale bars: b (low magnification), 50 µm; b (high magnification), 10 µm.
Figure S4 (related to Fig. 3): Double birthdating of donor neurons prior to transplantation identifies the stage and origin of cells undergoing successful cortical integration. (a) Recipient cortex after transplantation and retrograde labeling from the cp. The eGFP+ transplanted neurons (boxed areas in low magnification panels) and recipient SCPN are retrogradely labeled with CTB555 from the cp. There are three additional eGFP+ birthdated, transplanted, but retrograde CTB555 negative neurons (inset in high magnification CldU panel, top row, displays the CldU signal) in deeper layers of the same displayed section. These three neurons were double-positive for IdU and CldU, demonstrating that both birthdating and ICC were successful, and indicating that these three neurons were likely to be still mitotic or only very early postmitotic (less than 16 hours) at the time of transplantation. Analysis of eGFP+ and CTB555+ transplanted neurons shows that they are positive for IdU, but not CldU (arrows), demonstrating that these neurons were postmitotic for more than 16 hours before transplantation. The position of the boxes in the low magnification panels corresponds to the cortical area examined in the high magnification panels. eGFP (green), CTB555 (red), IdU (blue), CldU (purple). (b) Quantification from transplanted neurons analyzed with combined double IdU/CldU birthdating plus retrograde labeling (n=4). Double birthdating data are presented for three transplanted and retrogradely labeled SCPN (IdU+/CldU−; green in graphical representation); primary data from one additional neuron (shown here in black) is presented in Fig. 3. All analyzed neurons were IdU+/CldU−. These data indicate that early postmitotic neurons, encompassing already fate-restricted deep layer projection neurons and/or plastic post-mitotic neuroblasts with partially fate-restricted potential, account for the predominant population of neurons capable of achieving optimal level integration, and establishing subcerebral axon projections after transplantation.
Figure S5 (related to Fig. 4): Newly incorporated neurons of combinatorial subcerebral molecular identity establish anatomical subcerebral output connectivity with high fidelity. (a) High power confocal images of 3 eGFP+ (green) transplanted neurons retrogradely labeled with LumaFluor microspheres or CTB555 (red) from the cerebral peduncle demonstrate SCPN identity by high-level CTIP2 (purple) expression (as opposed to low-level expression, a characteristic of corticothalamic projection neurons) and by only barely detectable or completely absent callosal projection neuron identifier SATB2 (blue). Insets display retrogradely labeled layer V recipient SCPN within the same sections, and their respective CTIP2 (purple) expression levels for comparison. (b) High power confocal images of 3 additional eGFP+ (green) transplanted neurons retrogradely labeled with CTB555 (red) from the cerebral peduncle confirm SCPN identity and exclude identity of closely related corticofugal projection neuron populations by being negative for the corticothalamic identifier FOG2 (purple). DAPI (blue). Scale bars: a, b, 10 µm. (c) Quantification from transplanted neurons analyzed with combined molecular projection neuron subtype identity markers plus retrograde labeling (n=13). Molecular identity data are presented for six transplanted and retrogradely labeled SCPN (green in graphical representation); primary data from seven additional neurons (shown here in black) are presented in Figs. 4 and 6; Figs. S6, 8, and 12. Each row of dots matches with the same row molecular descriptions to their right. Taken together, these data reveal a remarkable level of molecular-to-projection fidelity, even compared with the very closely related corticofugal CThPN, since neither SATB2+ nor FOG2+ transplanted neurons projected subcerebrally.
Figure S6 (related to Fig. 4): Combined epifluorescence and confocal CTIP2 expression level analysis confirms SCPN identity of transplanted and subcerebrally projecting neurons. Analysis and comparison of CTIP2 expression levels (by epifluorescence and confocal microscopy) of CTB555 (red) retrogradely labeled recipient layer V SCPN and an eGFP+ (green) transplanted and subcerebrally projecting neuron, being localized in close vicinity at the border between layers IV and V. (a) Epifluorescence montage of upper cortical layers and adjacent layer V. The eGFP+ transplanted neuron and a recipient SCPN (indicated by arrow) with their respective nuclei are boxed and magnified in (b, c and g, h). Imaging was performed at the same exposure settings, demonstrating comparable high-level CTIP2 expression, in contrast to low-level CTIP2 expression as typical for related CFuPN populations including CThPN. (d – f) Single optical confocal section of the eGFP+ (green) transplanted neuron (boxed in (a)), unequivocally demonstrating that the analyzed high-level CTIP2 positive nucleus (purple) belongs to the transplanted and CTB555 (red) retrogradely labeled neuron. Scale bars: a, 50 µm; b – h, 10 µm. (i) Quantification from transplanted neurons analyzed with combined molecular projection neuron subtype identity markers plus retrograde labeling (n=13). Molecular identity data are presented for one transplanted and retrogradely labeled SCPN (green in graphical representation); primary data from 12 additional neurons (shown here in black) are presented in Figs. 4 and 6; Figs. S5, 8, and 12. Each row of dots matches with the same row molecular descriptions to their right. Taken together, these data indicate specific SCPN, rather than broad corticofugal projection neuron (CFuPN), identity.
Figure S7 (related to Fig. 5): Newly incorporated neurons of combinatorial callosal molecular identity establish anatomic inter-hemispheric output connectivity with high fidelity. (a) High power confocal images of 3 eGFP+ (green) transplanted neurons retrogradely labeled with CTB555 (red) from the contralateral hemisphere demonstrate CPN identity by expression of the CPN identifier SATB2 (blue) and being negative for or only exhibiting very low-level expression of CTIP2 (purple). (b) High power confocal images of 3 additional eGFP+ (green) transplanted and with CTB555 (red) from the contralateral hemisphere retrogradely labeled neurons further confirm CPN identity by SATB2 (blue) expression and being negative for FOG2 (purple). Scale bars: a, b, 10 µm. (c) Quantification from transplanted neurons analyzed with combined molecular projection neuron subtype identity markers plus retrograde labeling (n=9). Molecular identity data are presented for six transplanted and retrogradely labeled CPN (green in graphical representation); primary data from three additional neurons (shown here in black) are presented in Fig. 5. Each row of dots matches with the same row molecular descriptions to their right. Taken together, all analyzed transplanted and trans-callosally projecting neurons possess CPN molecular identity, and no transplanted neurons of SCPN or CThPN molecular identity were retrogradely labeled from the contralateral hemisphere.
Figure S8: Transplanted primed immature neurons rebuild subcerebral axon projections in Fezf2 knock-out recipient cortex. (a) Micro-transplantation of eGFP+ single cell suspension into Fezf2−/− P0/P1 recipient sensorimotor cortex. Ultrasound-guided injection of retrograde axonal tracer (CTB555) into the cerebral peduncle (cp) at P6. Analysis at P8. (b) Epifluorescence montage of a sagittal section of recipient Fezf2−/− cortex, including the adjacent part of the striatum (indicated by asterisk). There are 3 eGFP+ transplanted neurons in cortical layer VI (boxed area in in layer VI). CTB555 retrogradely labels the center neuron (arrow) from the cerebral peduncle (cp) and medium spiny neurons (MSNs; asterisk) in the striatum (due to the anatomical close proximity of the cp and MSN projection targets in the substantia nigra). Note the absence of retrograde label and high-level CTIP2 expression in layer V, demonstrating the absence of endogenous layer V SCPN in the Fezf2−/− recipient cortex (for comparison: the 3-ways split inset in the right upper corner displays control cortex after retrograde labeling from the cp; note the high-level CTIP2 staining and CTB555 labeling of layer V neurons (SCPN) and striatal MSN (asterisk)). Retrograde label of MSN in the Fezf2−/− recipient ascertains correct targeting of the cp with CTB555. The position of the box in layer VI corresponds to the cortical area examined in the adjacent confocal panel (first panel, top row; merge: eGFP, CTB555, CTIP2, SATB2) and to the inset in the right upper corner of the confocal panel (CTIP2 only) demonstrating high-level CTIP2 expression of the transplanted and retrogradely labeled neuron (arrow) in comparison to adjacent recipient-derived layer VI neurons. The position of the box in the confocal panel (first panel, top row) corresponds to the cortical area examined in high magnification panels, indicating high-level CTIP2 expression and absence of SATB2 expression in the retrogradely labeled transplanted neuron (arrow) (note: one of the two flanking (not retrogradely labeled) transplanted neurons also exhibits high-level CTIP2 expression, while the other one is negative for CTIP2). eGFP (green), CTB555 (red), SATB2 (blue), CTIP2 (purple). Scale bars: b (epifluorescence montage and first confocal panel, top row), 50 µm; b (high magnification confocal panels), 10 µm. (c) Quantification from transplanted neurons analyzed with combined molecular projection neuron subtype identity markers plus retrograde labeling (n=13). Molecular identity data are presented for one retrogradely labeled SCPN after transplantation into Fezf2−/− recipient cortex (green in graphical representation); primary data from 12 additional neurons (shown here in black) are presented in Figs. 4 and 6; Figs. S5, 6, and 12. Each row of dots matches with the same row molecular descriptions to their right. Taken together, these data not only reveal a remarkable level of molecular-to-projection fidelity, but, quite strikingly, demonstrate that newly incorporated (transplanted) neurons of SCPN molecular subtype identity extend subcerebral axon projections even in the absence of the endogenous pyramidal tract, thereby reconstituting otherwise entirely absent long-distance connectivity in the recipient brain.
Figure S9: Spines of transplantated neurons are juxtaposed to recipient-derived presynaptic synaptophysin suggesting synaptic integration with the recipient cortex. (a) Confocal stack 3D reconstructions of a transplanted eGFP+ neuron surrounded by synaptophysin-positive recipient-derived presynaptic terminals. (b) Epifluorescence image of the transplanted neuron. The boxed areas are magnified on the right (merge 3D reconstruction of a confocal stack; and high power magnification of single optical sections), showing a synaptophysin-positive recipient-derived presynaptic terminal flanked from two sides by eGFP+ synaptic spines on the apical dendrite of the transplanted neuron (indicated by arrows). These data are highly suggestive of synaptic integration of the transplanted neuron into the recipient cortex. eGFP (green), synaptophysin (red). Scale bars: a, 20 µm; b (epifluorescence image), 20 µm; b (merge 3D reconstruction), 10 µm; b (high power single optical sections), 10 µm.
Supplementary Figure 10 (related to Fig. 7 and Fig. 8): Channelrhodopsin-2-Venus expression in recipient-derived neurons or donor neurons renders neurons light sensitive. (a) tdTomato is used to visualize in utero-electroporated layer II/III neurons by epifluorescence in an acute slice preparation without exciting co-electroporated channelrhodopsin-2-Venus (gray scale panel). One neuron is patched (indicated by the arrows) and subsequently photo-stimulated with a blue LED. A biocytin-containing internal solution allowed for post hoc ICC analysis. A low power collapsed confocal stack (first panel) gives an overview of in utero-electroporated recipient-derived neurons in layers II/III (td-tomato+ and channelrhodopsin-2-Venus+). The position of the box corresponds to the cortical area examined in the high magnification panels giving a detailed view of the biocytin filled neuron. Note this neuron is positive for biocytin, cytoplasmic tdTomato and membrane-bound channelrhodopsin-2-Venus. Representative traces are shown on the right. Blue bars indicate light pulses. Top trace: Each light pulse robustly elicits a series of action potentials, thereby demonstrating sufficient expression of channelrhodopsin-2-Venus by in utero-electroporated neurons. Bottom trace: Corresponding action currents mediated by channlerhodopsin-2-Venus. The arrow points out an action current at a smaller time scale. Channelrhodopsin-2-Venus (green, membrane bound), tdTomato (red, cytoplasmic), biocytin (blue, cytoplasmic). Scale bars: a, 50 µm. (b, c) Two examples of patch clamp recordings of transplanted and channelrhodopsin-2-Venus and tdTomato in vitro Amaxa electroporated neurons are shown; transplanted neurons are visualized by epifluorescence of tdTomato in the acute slice preparation (gray scale panels). Depolarization of the transplanted neurons by current injection through the patch pipette elicits action potential trains, thereby confirming neuronal identity (middle panels). Photo-stimulation (blue bars) results in a robust depolarization and elicits several action potential spikes, thereby demonstrating sufficient expression of channelrhodopsin-2-Venus by in vitro Amaxa electroporated and transplanted neurons (right panels; top traces). Corresponding channelrhodopsin-2-Venus mediated action currents (right panels; bottom traces). Arrows point out action currents at a smaller time scale. Scale bars: b, c, 50 µm.
Figure S11: Rare cell fusion events do not account for the observed results. Transplantations of either control eGFP+/Fezf2+/− neuronal suspensions (contain all projection neuron subtypes) or eGFP+/Fezf2−/− suspensions (contain all projection neuron subtypes but SCPN, which were genetically removed from the mix) were performed and the axon projection pattern was analyzed. While transplanted Fezf2+/− neurons (a) send axons to corticofugal/subcortical targets (b [striatum], c [thalamus], d [tectum], e [pons]), Fezf2−/− neurons (f) reach (g) the striatum but fail to innervate more distal subcortical targets including (h) thalamus, (i) tectum and (j) pons (arguing against potential cell fusion events between transplanted and recipient derived SCPN, in which case a similar axon projection pattern would have been postulated). The inset in the top right corner in (f) demonstrates that Fezf2−/− neurons are capable of sending trans-callosal axons (the hemisphere contralateral to the transplanted cortex is shown on the right side; the boxed area is magnified in the left part of the inset, visualizing eGFP+ axons projecting from the corpus callosum up into the cortex). eGFP (green), L1 (red), NeuN (blue). Scale bars: a, f, 1000 µm; b – e and g – j, 250 µm; inset (right panel), 1000 µm; inset (left panel), 100 µm.
Figure S12: Rare cell fusion events do not account for the observed results. (a) Micro-transplantation of eGFP+ single cell suspension into P0/P1 Fezf2+/LacZ recipient sensorimotor cortex. Ultrasound-guided injection of retrograde axonal tracer (CTB555) into the cerebral peduncle (cp) at P6. Five-channel confocal (with spectral unmixing) analysis at P8. (b) The eGFP+ transplanted neuron (arrow) in layer V and adjacent recipient SCPN (arrowheads) in the same field of view are retrogradely labeled with CTB555 from the cerebral peduncle and express comparable high levels of CTIP2; the transplanted neuron is negative for Satb2 and Fezf2−b-gal while a large number of recipient SCPN in the same optical plane are Fezf2-b-gal positive. The position of the boxes corresponds to the cortical area examined in high magnification panels. The position of the inset boxes in the low magnification panels is magnified and shown as an inset in the high magnification Fezf2-b-gal panel for comparison. Fezf2-b-gal negativity of the transplanted neuron in comparison to positivity of recipient-derived neurons argues against fusion of eGFP+ transplant-derived cells with recipient-derived SCPN. eGFP (green), CTB555 (red), SATB2 (blue), CTIP2 (cyan), Fezf2-b-gal (purple). Scale bars: b (low magnification), 50 µm; b (high magnification), 10 µm; b (inset in the high magnification Fezf2-b-gal panel), 10 µm. (c) Quantification from transplanted neurons analyzed with combined molecular projection neuron subtype identity markers plus retrograde labeling (n=13). Molecular identity data are presented for one transplanted and retrogradely labeled SCPN (green in graphical representation); primary data from 12 additional neurons (shown here in black) are presented in Figs. 4 and 6; Figs. S5, 6, and 8. Each row of dots matches with the same row molecular descriptions to their right. Taken together, these data reveal a remarkable level of molecular-to-projection fidelity, even compared with the very closely related corticofugal CThPN, since neither SATB2+ nor FOG2+ transplanted neurons projected subcerebrally.
Figure S13: Quantification of molecular identity data. For analysis of molecular identity (CTIP2, SATB2, FOG2 ICC) combined with retrograde labeling (from cerebral peduncle or contralateral hemisphere), all neurons identified throughout all experiments and complying with the a priori criteria (neurons located in cortical gray matter separated from white matter by at least one layer of recipient-derived cortical neurons, and retrogradely labeled from target sites) were included in analysis; no neurons meeting these criteria were excluded. Representative neurons are depicted in the main figures (Figures 4, 5, 6), and all additional analyzed neurons in the Supplementary Figures (Figures S5, 6, 7, 8, 12). Each of these neurons is represented by a solid dot in the graphical representation. In total, n=13 transplanted neurons projecting to subcerebral targets (CSMN/SCPN) and n=9 neurons projecting to the contralateral hemisphere (CPN) (projection target is indicated at the bottom) were analyzed, and all display appropriate molecular identity for SCPN or CPN, respectively (molecular identity are indicated on the right side). Each row of dots matches with the same row molecular descriptions to their right. No transplanted and retrogradely labeled neurons were identified to have mismatching projection target and molecular identity (indicated by Ø). Fisher’s exact test was applied for statistical analysis, comparing the frequency of expression of a marker combination (SCPN or CPN) by neurons projecting to expected targets (cerebral peduncle or contralateral hemisphere) with the frequency of expression of a marker combination (SCPN or CPN) by neurons projecting to the alternative (mismatching) targets (contralateral hemisphere or cerebral peduncle): p<0.00001, demonstrating an extremely high level of molecular-to-projection fidelity. Fisher’s exact test is explicitly valid when a chi-squared test-based approximation is inadequate, since sample sizes are small, or the data are very unequally distributed among the cells of the table.
Rare cell fusion events do not account for the observed results.
First, transplantation into Fezf2−/− mice, with complete absence of endogenous SCPN and subcerebral pyramidal tract26,30,31, reveals that transplanted, newly incorporated neurons of SCPN molecular subtype identity extend subcerebral axon projections, reconstituting otherwise missing long-distance subcerebral connectivity in Fezf2−/− recipient brains (Fig. S8). Of further note, some transplanted neurons with subcerebral projections in wt recipients are localized slightly heterotopically, e.g. in layer VI (Fig. 4), which does not contain recipient SCPN. Cell fusion cannot account for the observed results.
Second, we performed the converse experiment, and transplanted eGFP+/Fezf2+/LacZ heterozygous (Fig. S11a) or eGFP+/Fezf2LacZ/LacZ null (Fig. S11f) donor neurons into wt recipients. Recipients were analyzed for eGFP+ axon projections in striatum, thalamus, contralateral cortex (Fezf2−/− mutant neurons are capable of projecting to all these tragets30,31), tectum, and pons (both of these subcerebral projections are absent in Fezf2−/− mice30,31). Corticofugal projections were analyzed one, two, and three weeks after transplantation, and trans-callosal projections were analyzed two weeks after transplantation. Strikingly, axon projections established by transplant-derived eGFP+/Fezf2LacZ/LacZ null neurons are only those formed endogenously by Fezf2−/− knockout neurons; while these transplant-derived Fezf2-null neurons project axons to striatum and contralateral cortex, they do not project axons to subcerebral regions, including tectum and pons (Fig. S11f – j and insets in f). In contrast, numerous eGFP+ axons of control transplant-derived eGFP+/Fezf2+/LacZ heterozygous neurons project to striatum, thalamus, and subcerebral regions (tectum, pons) (Fig. S11a – e), and these control heterozygous axons appear more organized and fasciculated in the striatum compared to more defasciculated axons from Fezf2-null transplant-derived neurons (as in Fezf2-null mice). These converse data more definitively rule out fusion, since this genetic deletion of the entire population of SCPN from the transplant source results in absence of subcerebral axon extension by transplant-derived neurons. These data also support the previous results on fidelity of molecular and projection identity, since the genetically Fezf2-null transplant-derived neurons maintain their limited range of subtype identity, and send axons only to targets appropriate for their genetic donor origin.
Finally, in Fezf2+/LacZ heterozygous mice, b-gal can serve both as a reporter for Fezf2 expression and as a marker to distinguish transplant-derived eGFP+ wt SCPN from recipient Fezf2 heterozygous, b-gal expressing SCPN. eGFP+ wt donor neurons were transplanted into Fezf2+/LacZ heterozygous recipients, and assessed by retrograde labeling from the cerebral peduncle at P6 (Fig. S12a). ICC for b-gal, CTIP2, and SATB2 demonstrates that endogenous SCPN, as expected, express high levels of CTIP2, are SATB2-negative, and are largely positive for b-gal (Fig. S12b). In contrast, transplanted wt eGFP+ neurons appropriately express high-level CTIP2, and are SATB2-negative, but importantly do not express b-gal (Fig. S12b, c). These results further reinforce that cell fusion is not responsible for the results reported here; neurons identified as transplant-derived wt eGFP+ neurons would have exhibited b-gal expression had fusion occurred. Together with the core labeling approaches used in all experiments, seven distinct, complementary modes of analysis rule out fusion as explaining the molecular and subtype fidelity, and bi-directional electrophysiological integration of developmentally primed transplanted neurons in these experiments.
Acknowledgments
This work was partially supported by grants to J.D.M. from the National Institutes of Health-NINDS NS041590 and NS049553, with additional infrastructure support from NS045523 and NS075672; from the ALS Association to J.D.M.; from The Regeneration Project to J.D.M., and from DFG grant Wu 590/2-1 to T.V.W.. F.M. was partially supported from a grant to V.N.M. (DC011291). H.P. was partially supported by an International Brain Research Organization (IBRO) Research Fellowship, and by a fellowship from The Regeneration Project. We thank P. Davis, D. Schuback, and Dr. K. Yee for superb technical assistance, and Dr. B. Goetze at the Harvard Center for Biological Imaging for excellent assistance with confocal spectral unmixing; Prof. M. Eichner at the Institute for Clinical Epidemiology and Applied Biometry at University of Tübingen for statistical advice; Drs. C. Lois, R. Hevner, M. Hibi, and S.K. McConnell for generous sharing of mice, antibodies, and reagents; and current and past members of the Macklis laboratory for helpful suggestions.
Footnotes
Author Contributions
T.V.W. and J.D.M. designed the overall experimental directions and specific analyses, and T.V.W., H.P., and J.D.M. wrote and edited the manuscript. T.V.W. also performed all non-electrophysiology-related experiments and data analysis, generated all experimental animals including those for electrophysiology, assisted with electrophysiology and performed post hoc immunocytochemical analysis of tissue sections used for electrophysiological evaluation. F.M. performed all electrophysiological recordings and data analysis and participated in manuscript editing. H.P. co-performed in utero-electroporation. A.P.W. assisted with mouse breeding and surgeries, immunocytochemistry, and microscopy. T.V.W., J.D.M., H.P., and V.N.M. contributed to data analysis and biological interpretation.
Competing financial interest (CFI) statement:
None of the authors have any competing financial interest (CFI) in this work.
References
- 1.Czupryn A, et al. Transplanted hypothalamic neurons restore leptin signaling and ameliorate obesity in db/db mice. Science. 2011;334:1133–1137. doi: 10.1126/science.1209870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Southwell DG, et al. Interneurons from embryonic development to cell-based therapy. Science. 2014;344:1240622. doi: 10.1126/science.1240622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baraban SC, et al. Reduction of seizures by transplantation of cortical GABAergic interneuron precursors into Kv1.1 mutant mice. Proc. Natl. Acad. Sci. USA. 2009;106:15472–15477. doi: 10.1073/pnas.0900141106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calcagnotto ME, et al. Effect of neuronal precursor cells derived from medial ganglionic eminence in an acute epileptic seizure model. Epilepsia. 2010;51:71–75. doi: 10.1111/j.1528-1167.2010.02614.x. [DOI] [PubMed] [Google Scholar]
- 5.Martínez-Cerdeño V, et al. Embryonic MGE precursor cells grafted into adult rat striatum integrate and ameliorate motor symptoms in 6-OHDA-lesioned rats. Cell Stem Cell. 2010;6:238–250. doi: 10.1016/j.stem.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Southwell DG, Froemke RC, Alvarez-Buylla A, Stryker MP, Gandhi SP. Cortical plasticity induced by inhibitory neuron transplantation. Science. 2010;327:1145–1148. doi: 10.1126/science.1183962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zipancic I, Calcagnotto ME, Piquer-Gil M, Mello LE, Alvarez-Dolado M. Transplant of GABAergic precursors restores hippocampal inhibitory function in a mouse model of seizure susceptibility. Cell Transplant. 2010;19:549–564. doi: 10.3727/096368910X491383. [DOI] [PubMed] [Google Scholar]
- 8.De la Cruz E, et al. Interneuron progenitors attenuate the power of acute focal ictal discharges. Neurotherapeutics. 2011;8:763–773. doi: 10.1007/s13311-011-0058-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka DH, Toriumi K, Kubo K, Nabeshima T, Nakajima K. GABAergic precursor transplantation into the prefrontal cortex prevents phencyclidine-induced cognitive deficits. J. Neurosci. 2011;31:14116–14125. doi: 10.1523/JNEUROSCI.2786-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunt RF, Girskis KM, Rubenstein JL, Alvarez-Buylla A, Baraban SC. GABA progenitors grafted into the adult epileptic brain control seizures and abnormal behavior. Nat. Neurosci. 2013;16:692–697. doi: 10.1038/nn.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stanfield BB, O'Leary DD. Fetal occipital cortical neurones transplanted to the rostral cortex can extend and maintain a pyramidal tract axon. Nature. 1985;313:135–137. doi: 10.1038/313135a0. [DOI] [PubMed] [Google Scholar]
- 12.Hernit-Grant CS, Macklis JD. Embryonic neurons transplanted to regions of targeted photolytic cell death in adult mouse somatosensory cortex re-form specific callosal projections. Exp. Neurol. 1996;139:131–142. doi: 10.1006/exnr.1996.0088. [DOI] [PubMed] [Google Scholar]
- 13.Shin JJ, et al. Transplanted neuroblasts differentiate appropriately into projection neurons with correct neurotransmitter and receptor phenotype in neocortex undergoing targeted projection neuron degeneration. J. Neurosci. 2000;20:7404–7416. doi: 10.1523/JNEUROSCI.20-19-07404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fricker-Gates RA, Shin JJ, Tai CC, Catapano LA, Macklis JD. Late-stage immature neocortical neurons reconstruct interhemispheric connections and form synaptic contacts with increased efficiency in adult mouse cortex undergoing targeted neurodegeneration. J. Neurosci. 2002;22:4045–4056. doi: 10.1523/JNEUROSCI.22-10-04045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magavi SS, Leavitt BR, Macklis JD. Induction of neurogenesis in the neocortex of adult mice. Nature. 2000;405:951–955. doi: 10.1038/35016083. [DOI] [PubMed] [Google Scholar]
- 16.Gaillard A, et al. Reestablishment of damaged adult motor pathways by grafted embryonic cortical neurons. Nat. Neurosci. 2007;10:1294–1299. doi: 10.1038/nn1970. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, Magavi SS, Macklis JD. Neurogenesis of corticospinal motor neurons extending spinal projections in adult mice. Proc. Natl. Acad. Sci. U S A. 2004;101:16357–16362. doi: 10.1073/pnas.0406795101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaspard N, et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008;455:351–357. doi: 10.1038/nature07287. [DOI] [PubMed] [Google Scholar]
- 19.Eiraku M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Ideguchi M, Palmer TD, Recht LD, Weimann JM. Murine embryonic stem cell-derived pyramidal neurons integrate into the cerebral cortex and appropriately project axons to subcortical targets. J. Neurosci. 2010;30:894–904. doi: 10.1523/JNEUROSCI.4318-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Espuny-Camacho I, et al. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron. 2013;77:440–456. doi: 10.1016/j.neuron.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Michelsen KA, et al. Area-specific reestablishment of damaged circuits in the adult cerebral cortex by cortical neurons derived from mouse embryonic stem cells. Neuron. 2015;85:982–997. doi: 10.1016/j.neuron.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Falkner S, et al. Transplanted embryonic neurons integrate into adult neocortical circuits. Nature. 2016;539:248–253. doi: 10.1038/nature20113. [DOI] [PubMed] [Google Scholar]
- 24.Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leyva-Díaz E, López-Bendito G. In and out from the cortex: development of major forebrain connections. Neuroscience. 2013;254:26–44. doi: 10.1016/j.neuroscience.2013.08.070. [DOI] [PubMed] [Google Scholar]
- 26.Arlotta P, et al. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 27.Alcamo EA, et al. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 28.Britanova O, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 29.Galazo MJ, Emsley JG, Macklis JD. Corticothalamic Projection Neuron Development beyond Subtype Specification: Fog2 and Intersectional Controls Regulate Intraclass Neuronal Diversity. Neuron. 2016;91:90–106. doi: 10.1016/j.neuron.2016.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 31.Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc. Natl. Acad. Sci. USA. 2005;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breunig JJ, Arellano JI, Macklis JD, Rakic P. Everything that glitters isn't gold: a critical review of postnatal neural precursor analyses. Cell Stem Cell. 2007;1:612–627. doi: 10.1016/j.stem.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 33.Alvarez-Dolado M, et al. Fusion of bone-marrow-derived cells with Purkinje neurons, cardio- myocytes and hepatocytes. Nature. 2003;425:968–973. doi: 10.1038/nature02069. [DOI] [PubMed] [Google Scholar]
- 34.Wagers AJ, Weissman IL. Plasticity of adult stem cells. Cell. 2004;116:639–648. doi: 10.1016/s0092-8674(04)00208-9. [DOI] [PubMed] [Google Scholar]
- 35.Goldman SA. Stem and Progenitor Cell-Based Therapy of the Central Nervous System: Hopes, Hype, and Wishful Thinking. Cell Stem Cell. 2016;18:174–188. doi: 10.1016/j.stem.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oki K, et al. Human-induced pluripotent stem cells form functional neurons and improve recovery after grafting in stroke-damaged brain. Stem Cells. 2012;30:1120–1133. doi: 10.1002/stem.1104. [DOI] [PubMed] [Google Scholar]
- 37.Tornero D, et al. Human induced pluripotent stem cell-derived cortical neurons integrate in stroke-injured cortex and improve functional recovery. Brain. 2013;136:3561–3577. doi: 10.1093/brain/awt278. [DOI] [PubMed] [Google Scholar]
- 38.Davies SJ, Field PM, Raisman G. Long interfascicular axon growth from embryonic neurons transplanted into adult myelinated tracts. J Neurosci. 1994;14:1596–1612. doi: 10.1523/JNEUROSCI.14-03-01596.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sadegh C, Macklis JD. Established monolayer differentiation of mouse embryonic stem cells generates heterogeneous neocortical-like neurons stalled at a stage equivalent to midcorticogenesis. J. Comp. Neurol. 2014;522:2691–2706. doi: 10.1002/cne.23576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sances S, et al. Modeling ALS with motor neurons derived from human induced pluripotent stem cells. Nat. Neurosci. 2016;16:542–553. doi: 10.1038/nn.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ladewig J, Koch P, Brüstle O. Auto-attraction of neural precursors and their neuronal progeny impairs neuronal migration. Nat. Neurosci. 2014;17:24–26. doi: 10.1038/nn.3583. [DOI] [PubMed] [Google Scholar]
- 42.Ziemba KS, Chaudhry N, Rabchevsky AG, Jin Y, Smith GM. Targeting axon growth from neuronal transplants along preformed guidance pathways in the adult CNS. J Neurosci. 2008;28:340–348. doi: 10.1523/JNEUROSCI.3819-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peron S, et al. A Delay between Motor Cortex Lesions and Neuronal Transplantation Enhances Graft Integration and Improves Repair and Recovery. J Neurosci. 2017;37:1820–1834. doi: 10.1523/JNEUROSCI.2936-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 45.Hirata T, et al. Zinc finger gene fez-like functions in the formation of subplate neurons and thalamocortical axons. Dev. Dyn. 2004;230:546–556. doi: 10.1002/dvdy.20068. [DOI] [PubMed] [Google Scholar]
- 46.Catapano LA, Arnold MW, Perez FA, Macklis JD. Specific neurotrophic factors support the survival of cortical projection neurons at distinct stages of development. J. Neurosci. 2001;21:8863–8872. doi: 10.1523/JNEUROSCI.21-22-08863.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molyneaux BJ, et al. DeCoN: genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron. 2015;85:275–288. doi: 10.1016/j.neuron.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vega CJ, Peterson DA. Stem cell proliferative history in tissue revealed by temporal halogenated thymidine analog discrimination. Nat Methods. 2005;2:167–169. doi: 10.1038/nmeth741. [DOI] [PubMed] [Google Scholar]
- 49.Petreanu L, Mao T, Sternson SM, Svoboda K. The subcellular organization of neocortical excitatory connections. Nature. 2009;457:1142–1145. doi: 10.1038/nature07709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev. Biol. 2001;240:237–246. doi: 10.1006/dbio.2001.0439. [DOI] [PubMed] [Google Scholar]
- 51.Franks KM, Isaacson JS. Synapse-specific downregulation of NMDA receptors by early experience: a critical period for plasticity of sensory input to olfactory cortex. Neuron. 2005;47:101–114. doi: 10.1016/j.neuron.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 52.Albeanu DF, Soucy E, Sato TF, Meister M, Murthy VN. LED arrays as cost effective and efficient light sources for widefield microscopy. PLoS One. 2008;3:e2146. doi: 10.1371/journal.pone.0002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Time course of subcerebral and trans-callosal axon extension by transplanted neurons. Transplanted neurons progressively extend long-distance axon projections and reach pons and more distal subcerebral targets as well as the contralateral cortex (f and g) by 5–6 days post-transplantation (dpt). Subcerebral axon projections: 4 dpt (a [striatum] and b [cerebral peduncle]), 5 dpt (c [pons] and d [proximal medulla oblongata]), 6 dpt (e [distal medulla oblongata]). Trans-callosal axon projections: 4 dpt (h), 5 dpt (i), 6 dpt (j). eGFP (green); L1 (red). Scale bars: a – e, 200 µm; h, – j, 100 µm.
Figure S2 (related to Fig. 2): Transplanted neurons assuming positions in cortical layers II–VI progressively mature and successfully extend trans-callosal long-distance axon projections. (a) Schematic illustrating the experimental outline. Micro-transplantation of eGFP+ single cell suspension into P0/P1 recipient sensorimotor cortex. Stereotaxic injection of retrograde axonal tracer (CTB555) into the contralateral hemisphere at P6. Analysis at P7 and P14. (b, c) A subset of transplanted neurons project trans-callosally. These neurons progressively mature from a DCX+ and NeuN−/low (b) to a DCX−/low and NeuN+ state (c). Compare the DCX expression of the recipient cortex at both time points. DCX decreases dramatically on a very similar time scale in the recipient cortex as for the transplanted neurons. The positions of the boxes in low magnification panels (b, c) correspond to the cortical area examined in high magnification panels. Arrows indicate transplanted neurons. eGFP (green), CTB555 (red), DCX (immature neuronal marker; purple), NeuN (mature neuronal marker; blue). Scale bars: b, c (low magnification), 50 µm; b, c (high magnification), 10 µm.
Figure S3 (related to Fig. 2): Long-distance axon projections of transplanted neurons remain stable long-term. (a) Schematic illustrating the experimental outline. Micro-transplantation of eGFP+ single cell suspension into P0/P1 recipient sensorimotor cortex. Stereotaxic injection of retrograde axonal tracer (CTB555) into the contralateral hemisphere at P35 and analysis at P37/P38. (b) A subset of transplanted neurons establish trans-callosal axon projections, remaining stable long-term, without pruning or retraction for periods as long as analyzed (5 weeks). These neurons progressively mature to a DCX− and NeuN+ state. The position of the box in the low magnification panel (b) corresponds to the cortical area examined in high magnification panels. Arrows indicate a transplanted neuron. eGFP (green), CTB555 (red), DCX (immature neuronal marker; purple), NeuN (mature neuronal marker; blue). Scale bars: b (low magnification), 50 µm; b (high magnification), 10 µm.
Figure S4 (related to Fig. 3): Double birthdating of donor neurons prior to transplantation identifies the stage and origin of cells undergoing successful cortical integration. (a) Recipient cortex after transplantation and retrograde labeling from the cp. The eGFP+ transplanted neurons (boxed areas in low magnification panels) and recipient SCPN are retrogradely labeled with CTB555 from the cp. There are three additional eGFP+ birthdated, transplanted, but retrograde CTB555 negative neurons (inset in high magnification CldU panel, top row, displays the CldU signal) in deeper layers of the same displayed section. These three neurons were double-positive for IdU and CldU, demonstrating that both birthdating and ICC were successful, and indicating that these three neurons were likely to be still mitotic or only very early postmitotic (less than 16 hours) at the time of transplantation. Analysis of eGFP+ and CTB555+ transplanted neurons shows that they are positive for IdU, but not CldU (arrows), demonstrating that these neurons were postmitotic for more than 16 hours before transplantation. The position of the boxes in the low magnification panels corresponds to the cortical area examined in the high magnification panels. eGFP (green), CTB555 (red), IdU (blue), CldU (purple). (b) Quantification from transplanted neurons analyzed with combined double IdU/CldU birthdating plus retrograde labeling (n=4). Double birthdating data are presented for three transplanted and retrogradely labeled SCPN (IdU+/CldU−; green in graphical representation); primary data from one additional neuron (shown here in black) is presented in Fig. 3. All analyzed neurons were IdU+/CldU−. These data indicate that early postmitotic neurons, encompassing already fate-restricted deep layer projection neurons and/or plastic post-mitotic neuroblasts with partially fate-restricted potential, account for the predominant population of neurons capable of achieving optimal level integration, and establishing subcerebral axon projections after transplantation.
Figure S5 (related to Fig. 4): Newly incorporated neurons of combinatorial subcerebral molecular identity establish anatomical subcerebral output connectivity with high fidelity. (a) High power confocal images of 3 eGFP+ (green) transplanted neurons retrogradely labeled with LumaFluor microspheres or CTB555 (red) from the cerebral peduncle demonstrate SCPN identity by high-level CTIP2 (purple) expression (as opposed to low-level expression, a characteristic of corticothalamic projection neurons) and by only barely detectable or completely absent callosal projection neuron identifier SATB2 (blue). Insets display retrogradely labeled layer V recipient SCPN within the same sections, and their respective CTIP2 (purple) expression levels for comparison. (b) High power confocal images of 3 additional eGFP+ (green) transplanted neurons retrogradely labeled with CTB555 (red) from the cerebral peduncle confirm SCPN identity and exclude identity of closely related corticofugal projection neuron populations by being negative for the corticothalamic identifier FOG2 (purple). DAPI (blue). Scale bars: a, b, 10 µm. (c) Quantification from transplanted neurons analyzed with combined molecular projection neuron subtype identity markers plus retrograde labeling (n=13). Molecular identity data are presented for six transplanted and retrogradely labeled SCPN (green in graphical representation); primary data from seven additional neurons (shown here in black) are presented in Figs. 4 and 6; Figs. S6, 8, and 12. Each row of dots matches with the same row molecular descriptions to their right. Taken together, these data reveal a remarkable level of molecular-to-projection fidelity, even compared with the very closely related corticofugal CThPN, since neither SATB2+ nor FOG2+ transplanted neurons projected subcerebrally.
Figure S6 (related to Fig. 4): Combined epifluorescence and confocal CTIP2 expression level analysis confirms SCPN identity of transplanted and subcerebrally projecting neurons. Analysis and comparison of CTIP2 expression levels (by epifluorescence and confocal microscopy) of CTB555 (red) retrogradely labeled recipient layer V SCPN and an eGFP+ (green) transplanted and subcerebrally projecting neuron, being localized in close vicinity at the border between layers IV and V. (a) Epifluorescence montage of upper cortical layers and adjacent layer V. The eGFP+ transplanted neuron and a recipient SCPN (indicated by arrow) with their respective nuclei are boxed and magnified in (b, c and g, h). Imaging was performed at the same exposure settings, demonstrating comparable high-level CTIP2 expression, in contrast to low-level CTIP2 expression as typical for related CFuPN populations including CThPN. (d – f) Single optical confocal section of the eGFP+ (green) transplanted neuron (boxed in (a)), unequivocally demonstrating that the analyzed high-level CTIP2 positive nucleus (purple) belongs to the transplanted and CTB555 (red) retrogradely labeled neuron. Scale bars: a, 50 µm; b – h, 10 µm. (i) Quantification from transplanted neurons analyzed with combined molecular projection neuron subtype identity markers plus retrograde labeling (n=13). Molecular identity data are presented for one transplanted and retrogradely labeled SCPN (green in graphical representation); primary data from 12 additional neurons (shown here in black) are presented in Figs. 4 and 6; Figs. S5, 8, and 12. Each row of dots matches with the same row molecular descriptions to their right. Taken together, these data indicate specific SCPN, rather than broad corticofugal projection neuron (CFuPN), identity.
Figure S7 (related to Fig. 5): Newly incorporated neurons of combinatorial callosal molecular identity establish anatomic inter-hemispheric output connectivity with high fidelity. (a) High power confocal images of 3 eGFP+ (green) transplanted neurons retrogradely labeled with CTB555 (red) from the contralateral hemisphere demonstrate CPN identity by expression of the CPN identifier SATB2 (blue) and being negative for or only exhibiting very low-level expression of CTIP2 (purple). (b) High power confocal images of 3 additional eGFP+ (green) transplanted and with CTB555 (red) from the contralateral hemisphere retrogradely labeled neurons further confirm CPN identity by SATB2 (blue) expression and being negative for FOG2 (purple). Scale bars: a, b, 10 µm. (c) Quantification from transplanted neurons analyzed with combined molecular projection neuron subtype identity markers plus retrograde labeling (n=9). Molecular identity data are presented for six transplanted and retrogradely labeled CPN (green in graphical representation); primary data from three additional neurons (shown here in black) are presented in Fig. 5. Each row of dots matches with the same row molecular descriptions to their right. Taken together, all analyzed transplanted and trans-callosally projecting neurons possess CPN molecular identity, and no transplanted neurons of SCPN or CThPN molecular identity were retrogradely labeled from the contralateral hemisphere.
Figure S8: Transplanted primed immature neurons rebuild subcerebral axon projections in Fezf2 knock-out recipient cortex. (a) Micro-transplantation of eGFP+ single cell suspension into Fezf2−/− P0/P1 recipient sensorimotor cortex. Ultrasound-guided injection of retrograde axonal tracer (CTB555) into the cerebral peduncle (cp) at P6. Analysis at P8. (b) Epifluorescence montage of a sagittal section of recipient Fezf2−/− cortex, including the adjacent part of the striatum (indicated by asterisk). There are 3 eGFP+ transplanted neurons in cortical layer VI (boxed area in in layer VI). CTB555 retrogradely labels the center neuron (arrow) from the cerebral peduncle (cp) and medium spiny neurons (MSNs; asterisk) in the striatum (due to the anatomical close proximity of the cp and MSN projection targets in the substantia nigra). Note the absence of retrograde label and high-level CTIP2 expression in layer V, demonstrating the absence of endogenous layer V SCPN in the Fezf2−/− recipient cortex (for comparison: the 3-ways split inset in the right upper corner displays control cortex after retrograde labeling from the cp; note the high-level CTIP2 staining and CTB555 labeling of layer V neurons (SCPN) and striatal MSN (asterisk)). Retrograde label of MSN in the Fezf2−/− recipient ascertains correct targeting of the cp with CTB555. The position of the box in layer VI corresponds to the cortical area examined in the adjacent confocal panel (first panel, top row; merge: eGFP, CTB555, CTIP2, SATB2) and to the inset in the right upper corner of the confocal panel (CTIP2 only) demonstrating high-level CTIP2 expression of the transplanted and retrogradely labeled neuron (arrow) in comparison to adjacent recipient-derived layer VI neurons. The position of the box in the confocal panel (first panel, top row) corresponds to the cortical area examined in high magnification panels, indicating high-level CTIP2 expression and absence of SATB2 expression in the retrogradely labeled transplanted neuron (arrow) (note: one of the two flanking (not retrogradely labeled) transplanted neurons also exhibits high-level CTIP2 expression, while the other one is negative for CTIP2). eGFP (green), CTB555 (red), SATB2 (blue), CTIP2 (purple). Scale bars: b (epifluorescence montage and first confocal panel, top row), 50 µm; b (high magnification confocal panels), 10 µm. (c) Quantification from transplanted neurons analyzed with combined molecular projection neuron subtype identity markers plus retrograde labeling (n=13). Molecular identity data are presented for one retrogradely labeled SCPN after transplantation into Fezf2−/− recipient cortex (green in graphical representation); primary data from 12 additional neurons (shown here in black) are presented in Figs. 4 and 6; Figs. S5, 6, and 12. Each row of dots matches with the same row molecular descriptions to their right. Taken together, these data not only reveal a remarkable level of molecular-to-projection fidelity, but, quite strikingly, demonstrate that newly incorporated (transplanted) neurons of SCPN molecular subtype identity extend subcerebral axon projections even in the absence of the endogenous pyramidal tract, thereby reconstituting otherwise entirely absent long-distance connectivity in the recipient brain.
Figure S9: Spines of transplantated neurons are juxtaposed to recipient-derived presynaptic synaptophysin suggesting synaptic integration with the recipient cortex. (a) Confocal stack 3D reconstructions of a transplanted eGFP+ neuron surrounded by synaptophysin-positive recipient-derived presynaptic terminals. (b) Epifluorescence image of the transplanted neuron. The boxed areas are magnified on the right (merge 3D reconstruction of a confocal stack; and high power magnification of single optical sections), showing a synaptophysin-positive recipient-derived presynaptic terminal flanked from two sides by eGFP+ synaptic spines on the apical dendrite of the transplanted neuron (indicated by arrows). These data are highly suggestive of synaptic integration of the transplanted neuron into the recipient cortex. eGFP (green), synaptophysin (red). Scale bars: a, 20 µm; b (epifluorescence image), 20 µm; b (merge 3D reconstruction), 10 µm; b (high power single optical sections), 10 µm.
Supplementary Figure 10 (related to Fig. 7 and Fig. 8): Channelrhodopsin-2-Venus expression in recipient-derived neurons or donor neurons renders neurons light sensitive. (a) tdTomato is used to visualize in utero-electroporated layer II/III neurons by epifluorescence in an acute slice preparation without exciting co-electroporated channelrhodopsin-2-Venus (gray scale panel). One neuron is patched (indicated by the arrows) and subsequently photo-stimulated with a blue LED. A biocytin-containing internal solution allowed for post hoc ICC analysis. A low power collapsed confocal stack (first panel) gives an overview of in utero-electroporated recipient-derived neurons in layers II/III (td-tomato+ and channelrhodopsin-2-Venus+). The position of the box corresponds to the cortical area examined in the high magnification panels giving a detailed view of the biocytin filled neuron. Note this neuron is positive for biocytin, cytoplasmic tdTomato and membrane-bound channelrhodopsin-2-Venus. Representative traces are shown on the right. Blue bars indicate light pulses. Top trace: Each light pulse robustly elicits a series of action potentials, thereby demonstrating sufficient expression of channelrhodopsin-2-Venus by in utero-electroporated neurons. Bottom trace: Corresponding action currents mediated by channlerhodopsin-2-Venus. The arrow points out an action current at a smaller time scale. Channelrhodopsin-2-Venus (green, membrane bound), tdTomato (red, cytoplasmic), biocytin (blue, cytoplasmic). Scale bars: a, 50 µm. (b, c) Two examples of patch clamp recordings of transplanted and channelrhodopsin-2-Venus and tdTomato in vitro Amaxa electroporated neurons are shown; transplanted neurons are visualized by epifluorescence of tdTomato in the acute slice preparation (gray scale panels). Depolarization of the transplanted neurons by current injection through the patch pipette elicits action potential trains, thereby confirming neuronal identity (middle panels). Photo-stimulation (blue bars) results in a robust depolarization and elicits several action potential spikes, thereby demonstrating sufficient expression of channelrhodopsin-2-Venus by in vitro Amaxa electroporated and transplanted neurons (right panels; top traces). Corresponding channelrhodopsin-2-Venus mediated action currents (right panels; bottom traces). Arrows point out action currents at a smaller time scale. Scale bars: b, c, 50 µm.
Figure S11: Rare cell fusion events do not account for the observed results. Transplantations of either control eGFP+/Fezf2+/− neuronal suspensions (contain all projection neuron subtypes) or eGFP+/Fezf2−/− suspensions (contain all projection neuron subtypes but SCPN, which were genetically removed from the mix) were performed and the axon projection pattern was analyzed. While transplanted Fezf2+/− neurons (a) send axons to corticofugal/subcortical targets (b [striatum], c [thalamus], d [tectum], e [pons]), Fezf2−/− neurons (f) reach (g) the striatum but fail to innervate more distal subcortical targets including (h) thalamus, (i) tectum and (j) pons (arguing against potential cell fusion events between transplanted and recipient derived SCPN, in which case a similar axon projection pattern would have been postulated). The inset in the top right corner in (f) demonstrates that Fezf2−/− neurons are capable of sending trans-callosal axons (the hemisphere contralateral to the transplanted cortex is shown on the right side; the boxed area is magnified in the left part of the inset, visualizing eGFP+ axons projecting from the corpus callosum up into the cortex). eGFP (green), L1 (red), NeuN (blue). Scale bars: a, f, 1000 µm; b – e and g – j, 250 µm; inset (right panel), 1000 µm; inset (left panel), 100 µm.
Figure S12: Rare cell fusion events do not account for the observed results. (a) Micro-transplantation of eGFP+ single cell suspension into P0/P1 Fezf2+/LacZ recipient sensorimotor cortex. Ultrasound-guided injection of retrograde axonal tracer (CTB555) into the cerebral peduncle (cp) at P6. Five-channel confocal (with spectral unmixing) analysis at P8. (b) The eGFP+ transplanted neuron (arrow) in layer V and adjacent recipient SCPN (arrowheads) in the same field of view are retrogradely labeled with CTB555 from the cerebral peduncle and express comparable high levels of CTIP2; the transplanted neuron is negative for Satb2 and Fezf2−b-gal while a large number of recipient SCPN in the same optical plane are Fezf2-b-gal positive. The position of the boxes corresponds to the cortical area examined in high magnification panels. The position of the inset boxes in the low magnification panels is magnified and shown as an inset in the high magnification Fezf2-b-gal panel for comparison. Fezf2-b-gal negativity of the transplanted neuron in comparison to positivity of recipient-derived neurons argues against fusion of eGFP+ transplant-derived cells with recipient-derived SCPN. eGFP (green), CTB555 (red), SATB2 (blue), CTIP2 (cyan), Fezf2-b-gal (purple). Scale bars: b (low magnification), 50 µm; b (high magnification), 10 µm; b (inset in the high magnification Fezf2-b-gal panel), 10 µm. (c) Quantification from transplanted neurons analyzed with combined molecular projection neuron subtype identity markers plus retrograde labeling (n=13). Molecular identity data are presented for one transplanted and retrogradely labeled SCPN (green in graphical representation); primary data from 12 additional neurons (shown here in black) are presented in Figs. 4 and 6; Figs. S5, 6, and 8. Each row of dots matches with the same row molecular descriptions to their right. Taken together, these data reveal a remarkable level of molecular-to-projection fidelity, even compared with the very closely related corticofugal CThPN, since neither SATB2+ nor FOG2+ transplanted neurons projected subcerebrally.
Figure S13: Quantification of molecular identity data. For analysis of molecular identity (CTIP2, SATB2, FOG2 ICC) combined with retrograde labeling (from cerebral peduncle or contralateral hemisphere), all neurons identified throughout all experiments and complying with the a priori criteria (neurons located in cortical gray matter separated from white matter by at least one layer of recipient-derived cortical neurons, and retrogradely labeled from target sites) were included in analysis; no neurons meeting these criteria were excluded. Representative neurons are depicted in the main figures (Figures 4, 5, 6), and all additional analyzed neurons in the Supplementary Figures (Figures S5, 6, 7, 8, 12). Each of these neurons is represented by a solid dot in the graphical representation. In total, n=13 transplanted neurons projecting to subcerebral targets (CSMN/SCPN) and n=9 neurons projecting to the contralateral hemisphere (CPN) (projection target is indicated at the bottom) were analyzed, and all display appropriate molecular identity for SCPN or CPN, respectively (molecular identity are indicated on the right side). Each row of dots matches with the same row molecular descriptions to their right. No transplanted and retrogradely labeled neurons were identified to have mismatching projection target and molecular identity (indicated by Ø). Fisher’s exact test was applied for statistical analysis, comparing the frequency of expression of a marker combination (SCPN or CPN) by neurons projecting to expected targets (cerebral peduncle or contralateral hemisphere) with the frequency of expression of a marker combination (SCPN or CPN) by neurons projecting to the alternative (mismatching) targets (contralateral hemisphere or cerebral peduncle): p<0.00001, demonstrating an extremely high level of molecular-to-projection fidelity. Fisher’s exact test is explicitly valid when a chi-squared test-based approximation is inadequate, since sample sizes are small, or the data are very unequally distributed among the cells of the table.
Data Availability Statement
All core data generated or analyzed during this study are included in this published article (and its supplementary information files). The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.