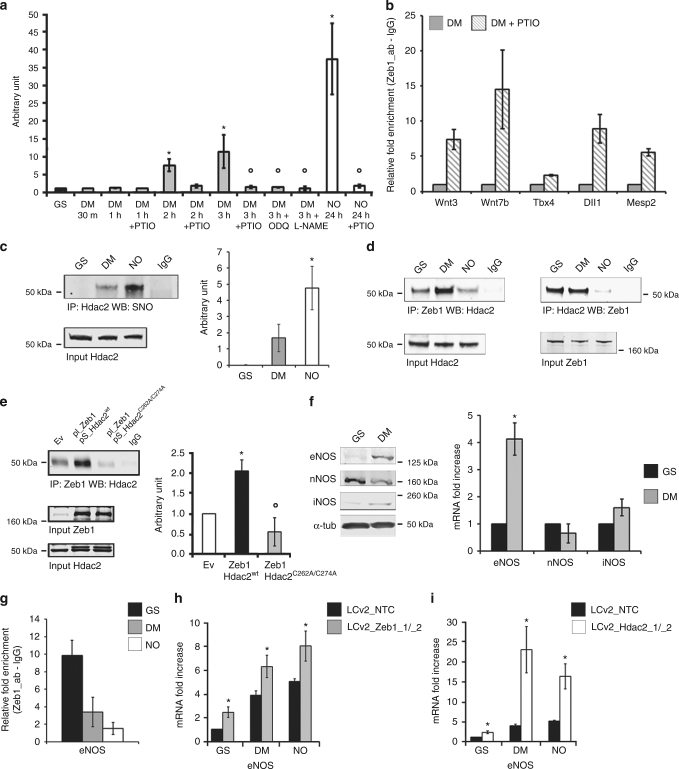
Fig. 3.
Endogenous NO synthesis and Zeb1-Hdac2 complex formation. a cGMP quantification in GS (black bar), in DM ± PTIO, ODQ, L-NAME (gray bars) for 1, 2, or 3 h and in NO ± PTIO (white bars) for 24 h. (n = 3; *p < 0.05 DM vs. GS; °p < 0.05 treatments vs. DM). b Zeb1-chromatin binding analysis on: Wnt3, Wnt7b, Tbx4, Dll1, and Mesp2. Chromatins extracted in DM (gray bars) or DM + PTIO (striped bars). Data compared to IgG value (striped bars) after Input normalization (n = 3). c Left panel: representative immunoprecipitation (IP)/western blotting (WB) analysis of Hdac2 S-nitrosylation in GS (24 h), DM or NO (2 h). Right panel: densitometry (n = 3; *p < 0.05 NO vs. DM). Full-length blot provided in Supplementary Fig. 9a. d Reciprocal co-IP analysis showing Zeb1:Hdac2 (left) and Hdac2:Zeb1 (right) complex formation in GS, DM or NO (24 h; n = 3 each group). Full-length blot provided in Supplementary Fig. 9b and c. e Left panel: representative co-IP analysis showing Zeb1:Hdac2wt and Zeb1:Hdac2C262A/C274A complex formation compared to empty vector (Ev)-transfected HeLa cells. HeLa transfected with Ev or co-transfected with pCMV6_Zeb1/pCIG2_Hdac2wt or pCMV6_Zeb1/pCIG2_Hdac2C262A/C274A. Right panel: densitometry (n = 3; *p < 0.05 Zeb1:Hdac2wt vs. Ev; °p < 0.05 Zeb1:Hdac2C262A/C274A vs. Zeb1:Hdac2wt). Full-length blot provided in Supplementary Fig. 9d. f Left panel: representative WB analysis of eNOS, nNOS, and iNOS in GS or DM (n = 3). Loading control: α-tubulin. Full-length blot provided in Supplementary Fig. 9e. Right panel: eNOS, nNOS, and iNOS mRNA analysis in GS (black bars) or DM (gray bars). Data expressed as fold increase of GS after subtraction of the housekeeping gene p0 signal (n = 3; *p < 0.05 DM vs. GS). g Zeb1-chromatin binding analysis on eNOS promoter in GS (black), DM (gray), and NO (white). Data represented as fold increase of GS after input normalization (n = 3). h, i eNOS mRNA expression analysis in GS, DM, and NO prior (control vector LCv2_NTC; black bar) and after CRISPR/Cas9 inactivation of Zeb1 (h; LCv2_Zeb1_1/_2; gray bars) or CRISPR/Cas9 inactivation of Hdac2 (i; LCv2_Hdac2_1/_2; white bar). Data represented as fold increase compared to control after subtraction of the housekeeping gene p0 signal (n = 3; *p < 0.05 LCv2_Zeb1_1/_2 or LCv2_Hdac2_1/_2 vs. LCv2_NTC). Data represented as mean ± s.e.m. and analyzed by Kolmogorov–Smirnov test