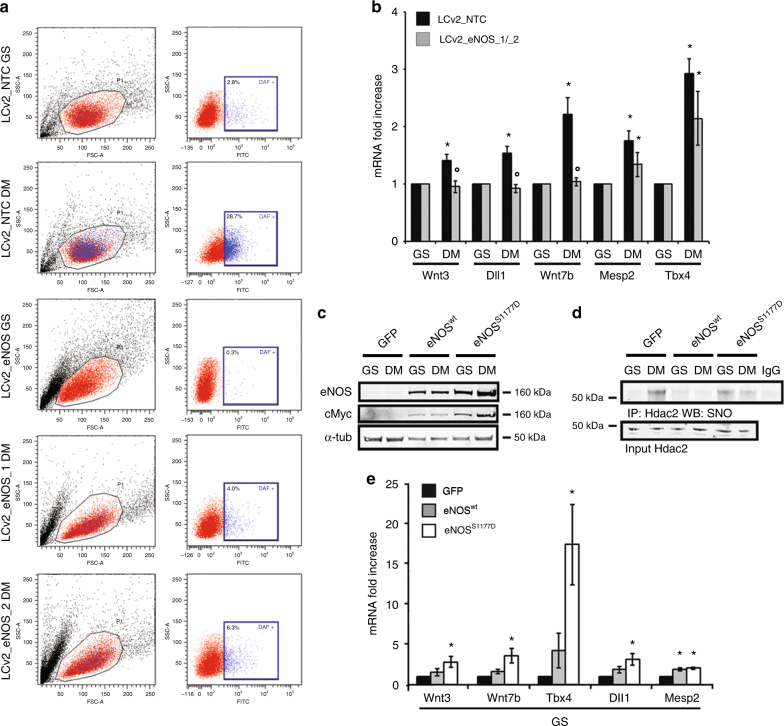
Fig. 5.
Role of eNOS during mesendodermal commitment. a Fluorescence activated cell sorter analysis of mESC cultured in GS and 2 h in DM prior (control vector LCv2_NTC) and after CRISPR/Cas9 inactivation of eNOS (LCv2_eNOS_1 or LCv2_eNOS_2). Left cytograms: representative scatter plots showing the forward (FSC-A) and side scatter (SSC-A) distribution of the mESC population. Right panels: representative dot plot showing the fluorescein isothiocyanate (FITC) and SSC-A distribution of the mESC population in the presence of DAF fluorescent probe. b qRT-PCR analysis of mRNA expression relative to Wnt3, Dll1, Wnt7b, Mesp2, and Tbx4 transcripts in mESC cultured for 24 h in GS and DM prior (control vector LCv2_NTC; black bar) and after CRISPR/Cas9 inactivation of eNOS (LCv2_eNOS_1/_2; gray bars). Data are shown as the mean of three independent experiments for each CRISPR/Cas9 vector ± s.e.m. represented as fold increase compared to control mESC after subtraction of the housekeeping gene p0 signal (*p < 0.05 DM vs. GS; °p < 0.05 LCv2_eNOS_1/_2 vs. LCv2_NTC). c Representative western blot performed with cell extracts obtained from mESC infected by Ad_GFP, Ad_eNOSwt, or Ad_eNOSS1177D cultured for 24 h in GS or DM (n = 3 each group). The expression of eNOS and cMyc was evaluated. The signal from α-tubulin antibody was used as loading control. Full-length blot provided in Supplementary Fig. 10a. d Representative immunoprecipitation (IP) western blotting (WB) analysis of Hdac2 S-nitrosylation performed in mESC infected by Ad_GFP, Ad_eNOSwt, or Ad_eNOSS1177D cultured in GS or after 2 h from release into DM (n = 3 each group). Full-length blot provided in Supplementary Fig. 10b. e qRT-PCR analysis of mRNA expression relative to Wnt3, Wnt7b, Tbx4, Dll1 and Mesp2 transcripts in mESC cultured for 24 h in GS prior (control Adeno virus Ad_GFP; black bar) and after infection by Ad_eNOSwt(gray bars) or Ad_eNOSS1177D expressing the constitutive active eNOS (white bars). Data shown as the mean of three independent experiments ± s.e.m. represented as fold increase compared to Ad-GFP infected mESC after subtraction of the housekeeping gene p0 signal (*p < 0.05 vs. Ad_GFP). Data analyzed by Kolmogorov–Smirnov test