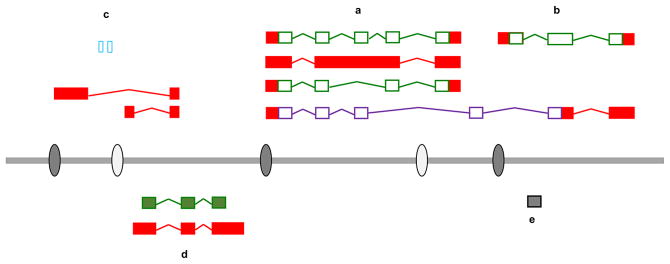
Figure 1. A modern view of the genomic landscape.
This hypothetical diagram illustrates the major types of genes and transcripts found in eukaryotic genomes. Two protein-coding genes are illustrated, (a) and (b). Coding sequences (CDS) are shown as open green boxes, untranslated regions as filled red boxes. Whereas locus (b) appears to generate a single CDS transcript, locus (a) generates two distinct protein isoforms through the differential incorporation of a central exon. Locus (a) also has a retained intron associated, while an additional ‘read-through’ transcript incorporates exons from (a) and (b). This transcript is subjected to NMD (unfilled lilac boxes). Gene (c) is a long non-coding RNA (lncRNA) with two transcripts (red boxes), although three small RNAs are also transcribed from within one of its introns (open blue boxes). Loci (d) and (e) are unprocessed (filled green boxes) and processed (grey box) pseudogenes respectively. Locus (d) is transcribed. A series of promoter regions (filled grey ovals) and enhancer regions (open ovals) are indicated. Promoters are associated with transcription start sites (TSSs) for the various loci, whereas enhancers are found some distance from the gene or genes they regulate.