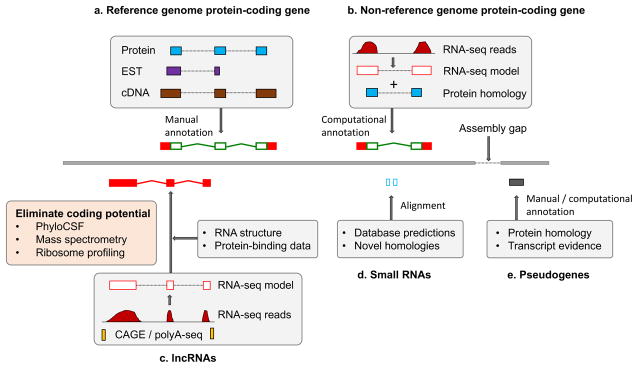
Figure 2. The core annotation workflows for different gene types.
These workflows illustrate general annotation principles, not the specific pipelines of any particular genebuild. a) Protein-coding genes within reference genomes were largely annotated based on the computational genomic alignment of Sanger-sequenced transcripts and protein-coding sequences, followed by manual annotation via interface tools such as Zmap1, WebApollo26, Artemis126 or the Integrative Genomics Viewer127. Transcripts were typically taken from GenBank128, proteins from Swiss-Prot12. b) Protein-coding genes within non-reference genomes are usually annotated based on fewer resources; here, RNA sequencing (RNA-seq) data are used in combination with protein homology information extrapolated from a closely-related genome. RNA-seq pipelines for read alignment include STAR129 and TopHat130, whereas model creation is commonly performed by Cufflinks22. c) Long non-coding RNA (lncRNA) structures can be annotated in a similar manner to protein-coding transcripts as for (a) and (b), although coding potential must be ruled out. This is typically done by examining sequence conservation with phyloCSF131 or using experimental datasets such as mass spectrometry or ribosome profiling. Here, 5’ Cap Analysis of Gene Expression (CAGE)45 and polyA-seq data46 are also incorporated to obtain true transcript endpoints. Designated lncRNA pipelines include PLAR48. d) Small RNAs are typically added to genebuilds by mining repositories such as RFAM132 or miRBase133. However, these entries can be used to search for additional loci based on homology. e) Pseudogene annotation is based on identification of loci with protein-homology to either paralogous or orthologous protein-coding genes. Computational annotation pipelines include PseudoPipe52, although manual annotation is more accurate53. Finally, all annotation methods can be thwarted by the existence of sequence gaps in the genome assembly (right-angled arrow).