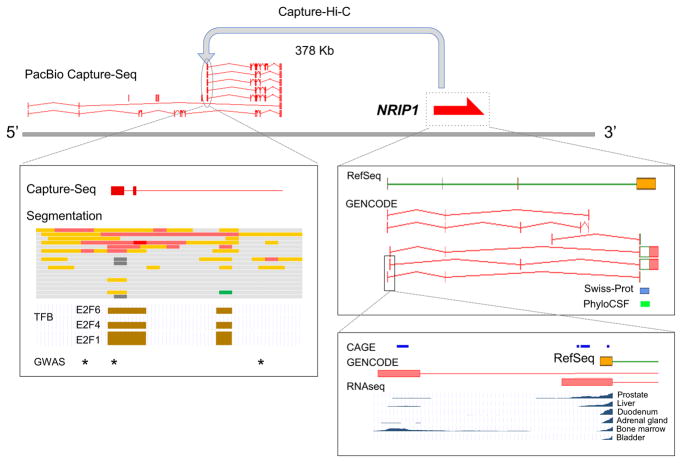
Figure 4. Transcriptional complexity in the NRIP1 locus.
a) Capture Hi-C105 indicates that the nuclear receptor interacting protein 1 (NRIP1) locus on human chromosome 21 forms a loop with a previously unannotated region nearby. Pacific Biosciences (PacBio) CaptureSeq data could be aligned here (R. Johnson, personal communication), leading to the annotation of lncRNA OTTHUMG00000488671 in GENCODE. b)| A long non-coding RNA (lncRNA) transcription start site (TSS) falls within an ENCODE-defined enhancer 102 (red and orange blocks; processed by Ensembl134). Three transcription factor binding (TFB) regions — E2F1, E2F4 and E2F6 — co-localize based on ENCODE chromatin immunoprecipitation followed by sequencing (ChIP-seq) data102. In combination, these data suggest an ‘extended gene model’ for NRIP1, which may aid the interpretation of three genome-wide association study (GWAS) signals linked to Crohn’s disease (rs2823286, rs1297265 and rs1736020; shown as asterisks) as previously noted by Mifsud et al.105 c) NRIP1 contains one transcript in RefSeq and 6 in GENCODE. The coding sequence (CDS; shown as an open green box) has Swiss-Prot support, and a PhyloCSF conservation signal131. (The untranslated regions (UTRs) are shown as filled red boxes.) d) Two distinct first exons of NRIP1 are annotated, both supported by 5’ Cap Analysis of Gene Expression (CAGE) data45. RNA-seq from Uhlen et al.115 indicates differential expression, with usage of the upstream exon apparently limited to bone marrow (and adipose; not shown). This TSS is dominant in white blood cells, which are bone-marrow-derived. RNA-seq and CAGE support a more general expression profile for the downstream first exon, with evidence of TSS variability.