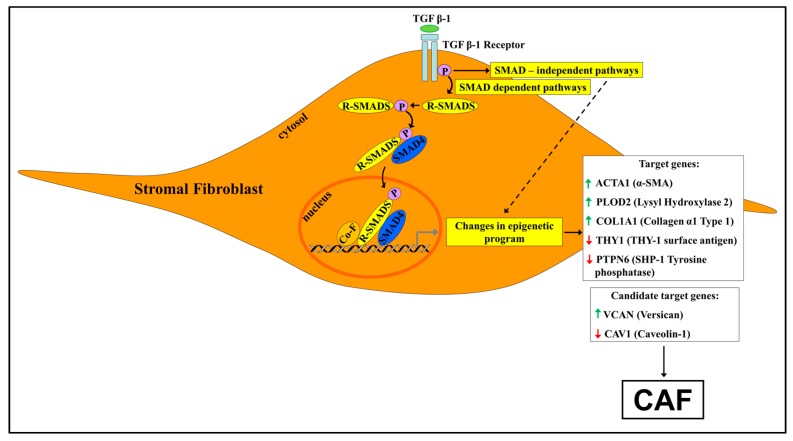
Figure 2.
Cancer-derived TGF-β1 reshapes the epigenetic signature of normal stromal fibroblasts surrounding a growing tumor via SMAD-dependent and SMAD-independent signaling pathways. For clarity’s sake, only the SMAD-dependent pathway is shown. TGF-β1 signaling induces changes in nuclear enzymes and regulatory proteins such as DNMTs, histone-modifying enzymes and BET proteins governing the multi-tiered epigenetic program of fibroblast cells. The outcome is chromatin reprogramming and the generation in the tumor microenvironment (TME) of pro-tumorigenic CAFs bearing a stable phenotype. CAFs, in turn, secrete TGF-β1 and therefore participate in a vicious pro-tumorigenic cycle shown in Figure 1. Target candidate genes denote stromal fibroblast genes known to be regulated by TGF-β for which, in contrast to target genes, experimental evidence for TGF-β reshaping of epigenetic landscape is, to date, circumstantial. This figure is based on selected papers discussed in the review presenting evidence for TGF-β1-induced alterations in the epigenetic signature of stromal fibroblasts during trans-differentiation to CAFs in cancer (target genes) or in myofibroblasts resident in non-neoplastic fibrotic loci (candidate genes). Green arrow: up-regulation. Red arrow: down-regulation. In brackets: the coded protein. Co-F: co-factors.