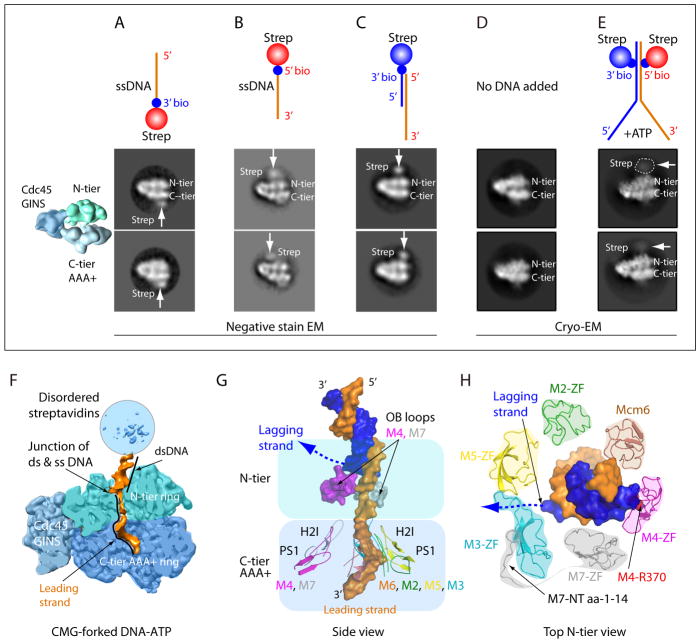
Figure 4. CMG-forked DNA structure with N-tier leading the C-tier during translocation.
A–C) Negative stain EM side views of CMG with streptavidin tipped oligos; colored diagram at left illustrates the tiers of Mcm2-7 and Cdc45/GINS. A) 20mer ssDNA 3′ streptavidin. B) 20mer ssDNA 5′ streptavidin. C) fork lacking a lagging strand and tipped with streptavidin on the ds end. D) CryoEM of CMG without DNA. Note the density of Mcm6 WHD protruding from the C-tier. E) CryoEM of CMG-forked DNA and ATP; CMG is stopped at the streptavidin blocks which are visible as a fuzzy (mobile) region. F) Cut away view through CMG-forked DNA illustrating dsDNA enters the top of the N-tier (light green) and leading ssDNA proceeds into the motor C-tier (blue). G) The DNA-interacting loops of Mcm2-7. Note that the upper OB loops of Mcm4 and Mcm7 block the lagging strand DNA. Inside the C-tier motor ring, the PS1 and H2I loops of Mcm 2, 3, 5, 6 spiral around and interact with the right-handed leading strand DNA coil; the corresponding loops in Mcm 4, 7 do not interact with DNA. Lagging ssDNA is not visible but a possible path is suggested by the dotted line. H) Top view of N-terminal ZF domains of Mcm2-7 binding the dsDNA. Note dsDNA tilts toward and interacts with ZFs of Mcm 7, 4, 6. The Mcm3 ZF has an unusually long loop that forms the base between ZFs of Mcm 3, 5. The ordered N-terminal peptide of Mcm7 warps around and pulls Mcm3 ZF from Mcm5 ZF, creating a gap through which the lagging strand may escape (dotted line). The EM images and illustrations in panels A- E are adapted with permission from Figs 5 and 6 of 23.