Figure 1. The C. jejuni Transcriptome in Human Infection and Chick Commensalism.
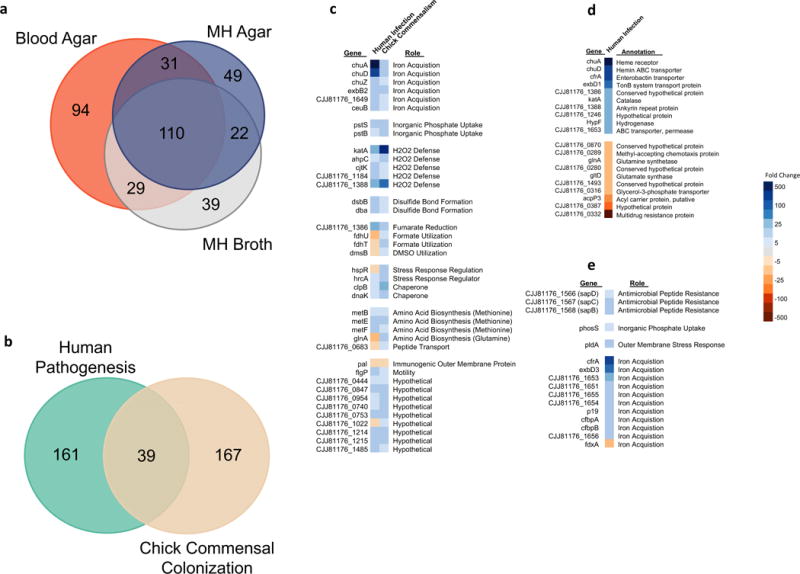
a, Differentially regulated genes in vivo. A Venn diagram showing the number of differentially regulated C. jejuni CG8421 genes (fold change >|3|, FDR p-value <0.05) in the infected feces of three volunteers compared to laboratory control growths which each represent three biological replicates. b, Human infection versus chicken commensalism transcriptomes. A Venn diagram showing the number of differentially regulated (fold change >|3|, FDR p-value <0.05) gene homologs in vivo between C. jejuni CG8421 infected human feces (3 samples) and a previously published RNA-seq transcriptome of C. jejuni 81-176 in the chicken cecum, which represents 3 pools of 5 infected chick ceca each. Both data sets use mid-log phase growth in Mueller-Hinton broth biological triplicates as the in vitro lab comparison. c, Conserved differential gene regulation across hosts, lifestyles, and strains. The 39 differentially regulated homologs from Fig. 1b conserved between human infection and chicken commensalism with transcriptional fold changes noted. d, The top most upregulated and downregulated CG8421 genes in human infection samples. e, Notable genes uniquely differentially regulated in human infection samples.
