Figure 4. Validating C. jejuni Genomic Variant Selection in non-Human Primates.
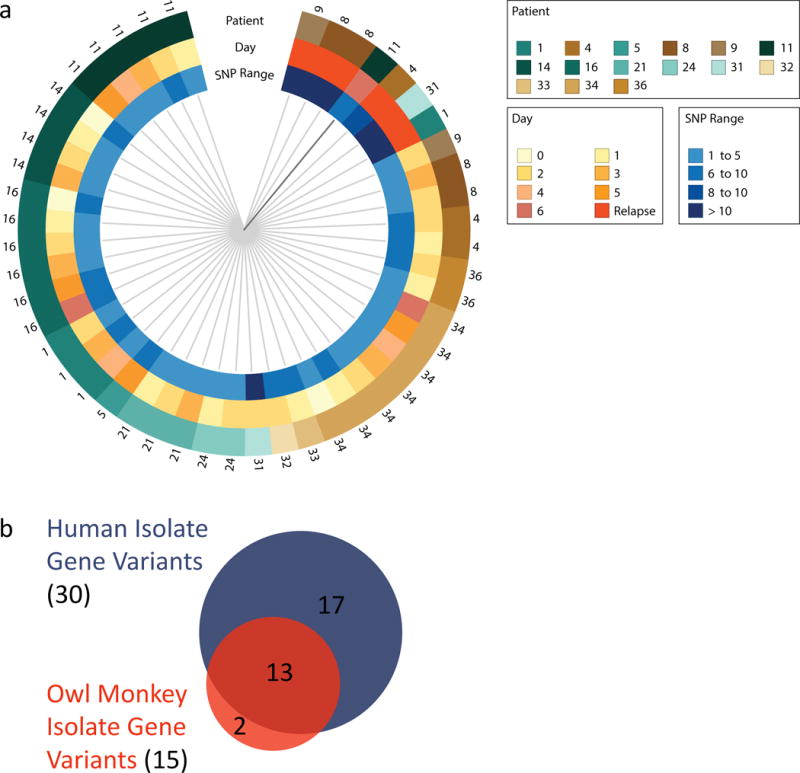
a, A SNP tree representing how similar human infection isolate populations are to one another. Each wedge represents one sequenced isolate population and the closer two samples are to each other the more similar their genomic variations. Volunteer identification numbers per sample are noted on the outside of the ring, and the day of infection and the number of SNPs detected are denoted by the inner rings. Note that primary infection isolates from the same volunteer are more similar to each other than they are to isolates from other volunteers. However, relapse infection isolates (bright orange) are more similar to each other despite the volunteer they come from. b, Conserved gene variant selection across primates. A second, different C. jejuni CG8421 inoculum was used to infect owl monkeys, and the genomes of 12 diarrheal primary infection isolate populations from 5 monkeys were sequenced (Sup. Tables 12 and 13). The Venn diagram shows the C. jejuni genome variants selected for in owl monkey infection closely resemble those of human infection (Sup. Table 13). This supports the human data and suggests non-human primates harbor a similar fitness selection environment found in humans. The smaller sample size and no relapse infections contributed to fewer total variants detected in the monkeys.
