Figure 1. Conservation of ZFP568 and its Igf2-P0 binding site in eutheria.
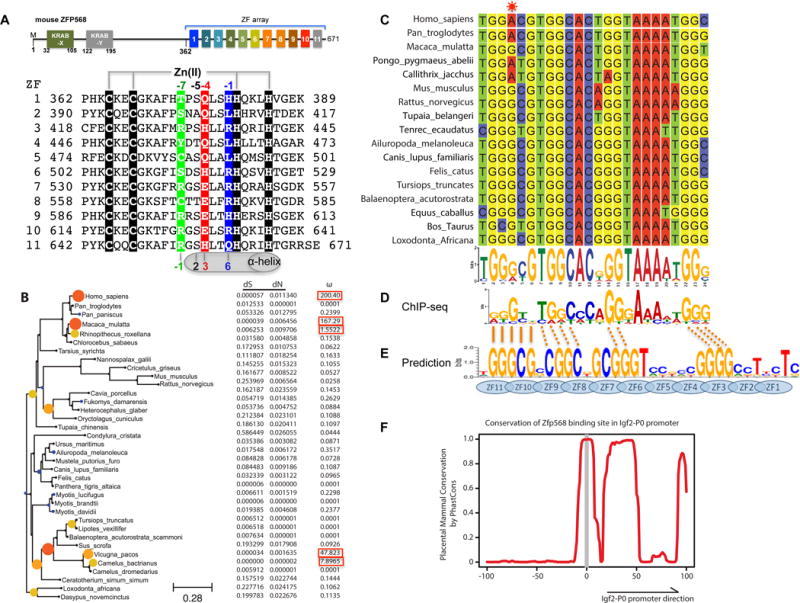
(A) Schematic representation of mouse ZFP568 and sequence alignment of its 11 zinc fingers. The four Zn-coordinating residues of each finger are highlighted with white letters against black. In conventional C2H2 ZF proteins, the amino acids occupying four key ‘canonical’ positions of the helix, namely −1, 2, 3 and 6, specify a DNA target sequence of three or four adjacent DNA base pairs (bottom). This structure-based numbering scheme refers to the position immediately prior to the helix (−1) and positions within the helix (2, 3 and 6). To reduce possible ambiguity, we use the first zinc-coordination His in each finger as reference position 0, with residues prior to this, at sequence positions −1 (blue), −4 (red), −5, and −7 (green), corresponding to the 6, 3, 2 and −1 of the structure-based numbering (compare top and bottom). The new numbering scheme (residues at positions −1, −4, and −7) corresponds to the 5′-middle-3′ of each DNA triplet element.
(B) Maximum likelihood phylogenetic tree and selection pressure for ZFP568 zinc finger domains in mammals. The ω (dN/dS) value for each branch is indicated by the color and size of the round disk on the node, with darker color and larger size for higher ω value. Human ZNF568 H allele was used for this analysis.
(C) Alignment of the Igf2-P0 locus from 17 representative placenta mammals.
(D) mZFP568 consensus binding motif as determined by ChIP-seq.
(E) Prediction made by polynomial SVM (support vector machines)-based algorithm.
(F) Plot of placental mammal conservation in a 100bp window flanking the Igf2-P0 site. See also Figure S1 and Table S1.
